Research - (2021) Volume 9, Issue 2
Age Estimation Using DNA Extracted from Human Saliva as a Diagnostic Measure in Forensic Odontology
Keerthana T and Sindhu Ramesh*
*Correspondence: Sindhu Ramesh, Department of Conservative Dentistry and Endodontics, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, India, Email:
Abstract
Forensic age estimation can be used to gain information relevant to criminal and anthropological investigations. Forensic dentistry involves the processing, review, evaluation and presentation of dental evidence with the purpose of contributing scientific and objective data in legal processes. This study aims in estimating age of a person using DNA extracted from human saliva and to assess this methodology as a potential diagnostic measure in forensic odontology. Salivary samples were collected from 45 blinded participants aged 18 to 70 years with a DNA self-collection kit and stored at room temperature. DNA was extracted from the salivary samples using a separate kit and it was subjected to age estimation process by constructing an age predictive model. A constructed age predictive model of saliva has 6 selected CpG sites (5’—C---phosphate—G---3’) in genome-enabled age prediction with high accuracy. Univariate linear regression analysis was performed to test the association between age and gender.(p value<0.05). DNA methylation profiling of saliva was performed to identify age associated CpG markers. A model composed of 6 selected CpG sites enabled age prediction in saliva with high accuracy and this multiplex system can be integrated into the routine forensic laboratory workflow after further validation tests with various casework samples.
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Keywords
Saliva, DNA, epigenetic age marker, Methylation, CpG sites
Introduction
Individual age estimation is a major key factor in forensic science analysis which can provide very useful information applicable to crime investigations. Age estimation in forensic science was initially based on various morphological approaches or radiographic investigations [1]. Later, molecular approaches came into practice for age estimation [2,3]. DNA based methodologies initially were technique sensitive and had lack of accuracy. But, epigenetic signature of DNA methylation had overcome these disadvantages. Most forensic age predictor models have been developed based on blood DNA samples, because it is the most encountered fluid in the crime scene. But now, additional tissues are now being explored [4,5]. Saliva is the next frequently encountered fluid in the crime scene. Based on salivary samples, epigenetic age predictive models were formed to predict the age of a suspected person in the crime scene.
DNA phenotyping refers to infer the appearance of an unknown sample donor from DNA [16]. When there is no suspect to match or DNA profile from the evidence does not match with anyone, forensic DNA phenotyping is expected to provide an investigate lead that can facilitate a traced search for an unknown suspect by narrowing the search range. The most well studied externally visible characteristics are those associated with the pigmentation such as skin color, eye color, and hair color [7,8]. But age is noteworthy because it can be used regardless of ethnicity to predict an individual’s appearance.
To predict an individual’s age from DNA, molecular methods based on the detection of telomere shortening and mitochondrial DNA deletion have been suggested [9,10]. DNA methylation based age prediction can be applied to various tissues, body fluids which has resulted in several researches reporting age associated DNA markers and age predictive models [11,12].
DNA methylation based biomarkers often referred to as epigenetic age or ‘epigenetic clock’ as they are robust estimators of chronological age of an individual [13,14]. This estimate is consistent across many biological specimens such as blood, brain, breast, kidney, liver, lung, saliva and cell types including CD4+ T cells, monocytes, glial cells and neurons [15,16]. Epigenetic age is associated with age related health outcomes above and beyond chronological age. Thus, epigenetic age captures some sort of biological age and the resulting susceptibility to the disease and multiple health outcomes.
Further, these age predictive models would not be practical to implement in routine forensic laboratories that usually deal with very limited amounts of DNA. Therefore, models composed of a small number of highly predictive CpG markers in a certain type of body fluid or tissue that have been reported to produce reasonable prediction accuracy on several different analysis platforms. Usually salivary samples have a heterogeneous cell composition that includes both buccal epithelial cells and leukocytes [8,17,18]. This study aims at generating genome wide DNA methylation profiles using bisulfite sequencing and assessing those age predictive models can be suitable for predicting the range of suspected individuals. This study also emphasizes whether using salivary DNA samples can be a potential diagnostic aid with certain prediction accuracy in forensic practice.
Materials and Methods
Study design
Single blinded in vivo study.
Ethical clearance
Ethical permission and approval for the project was obtained from the Institutional Review Board of Saveetha Institute of Medical and Technical Sciences, Chennai, India. (SBA/2019/23/05).
Study criteria
Salivary samples were collected from 45 volunteers aged 18 to 70 years with a DNA selfcollection kit and stored at room temperature and sent to the lab. DNA was extracted from the salivary samples using a separate kit and it was subjected to age estimation process by constructing an age predictive model.
Sample size
Salivary samples were collected from 45 volunteers among which 23 were males, and 22 were females in the age group between 18 to 60 years. The salivary samples were collected using a DNA self-collection kit and stored at room temperature.
Procedure
The steps involved are
✓ Sample collection.
✓ Genome wide DNA methylation profiling of saliva.
✓ Targeted bisulfite sequencing.
✓ Construction of age predictive model for saliva.
Sample collection
After the collection process, DNA was segregated from the stored salivary samples ranging from 150 to 200 microliters of saliva using DNA mini kit and then quantified.
The protocol followed before collecting saliva was to wait one hour after eating a meal before collecting saliva. Patients were asked to rinse their mouths thoroughly to remove any food particles or other contaminants. After 5 to 10 minutes the salivary samples were collected. Collecting saliva soon after rinsing may reduce the amount of DNA extraction and may also affect the biomarker analysis [19,20].
Salivary DNA self-collection kit provided the convenience of integrated ambient temperature stabilization, sample transport, and sample preparation. The device is available in several formats [21,22]. In this study, tube format was used, for that whole saliva should be delivered directly into the device by placing the kit against the lower border and spitting into the funnel shaped end.
The DNA in saliva originates from cells that are shed from the inner linings of the mouth and from white blood cells [23,24]. The quantification of samples was done by polymerase chain reaction. For quantification, as per manufacturer’s instructions, a mix solution of 10 microliters of powerquant* mastermix; 7 microlitres of amplification grade (water); 1 microliter of power-quant* primer mix. The quantification was made on 7500 real time PCR cycles.
Genome wide DNA methylation profiling of saliva
Microarray hybridization was performed. 1 to 2 micrograms of DNA was bisulfite converted using a DNA methylation kit according to the manufacturer's instructions. Bisulfite converted DNA was then incubated at 37 degrees for 24 hours. The amplified DNA was then fragmented and hybridized to a hybridization oven at 48 degrees for 12 to 18 hours.
Following hybridization, single base extension of hybridized DNA was performed using hapten modified nucleotides [25,26]. Staining was then performed by adding fluorescently labeled antibodies in several steps to amplify the signal, the arrays were washed.
Data collection
Then, univariate linear regression analysis was performed to test the association between beta score and age separately for each CpG site. The CpG sites that satisfied the following criteria were selected as age-associated CpG marker candidates: false discovery rate-adjusted p-value<0.05, and difference between maximum and minimum beta scores >0.5 (Table 1 and 2).
| Variable | Regression coefficient | p value |
|---|---|---|
| Age | 0.238 | 0.02 |
| Gender | 0.332 | 0.03 |
Table 1: Showing univariate linear regression analysis among the variables.
| CpG sites | Location in Genes | Methylation levels |
|---|---|---|
| cg12570511 | TBR1 | 0.34 |
| cg 07547549 | SLC12A5 | 0.26 |
| cg 18384097 | PTPN7 | 0.41 |
| cg00481951 | SST | 0.37 |
| cg14361627 | KLF14 | 0.24 |
| cg08428415 | TSSK6 | 0.32 |
Table 2: Showing methylation levels of CpGs in multiplex methylation assays.
Targeted bisulfite sequencing using methylation snapshot
Selected CpG markers were further investigated using multiplex SNaPshot reaction of bisulfiteconverted DNA. In silico bisulfite-converted genomic reference sequences determined from the BeadChip results were targeted in bisulfite sequencing [27,28]. PCR primers for the amplification of bisulfite converted genomic DNA were designed using the MethPrimerprogram PyroMark1 Assay Design software version 2.0 (Qiagen), and single-base extension (SBE) primers for target CpGs within the PCR products were designed using the BatchPrimer3 program PCR was performed in 20mL reaction volumes containing 1 to 2mL of bisulfite-converted DNA, 3 of AmpliTaq Gold 1 DNA Polymerase (Applied Biosystems), 2mL of Gold ST*R 10 Buffer (Promega, Madison, WI, USA), and 0.2–3.0mM of each primer.Bisulfite-converted DNA was obtained by modification of 500 ng of genomic DNA using the EpiTect1 Fast DNA Bisulfite Kit (Qiagen) or by modification of less than 10 ng of genomic DNA with the Imprint 1 DNA Modification Kit according to the manufacturers’ protocols.
PCR cycling was conducted using a VeritiTM 96-Well Thermal Cycler under the following conditions: 95°C for 11 min; 34 cycles of 94°C for 20 s,56°C for 60 s, and 72°C for 30 s; and a final extension at 72°C for 7 min. Then, 5mL aliquots of PCR products were purified with 1mL of ExoSAP-ITTM by incubation at 37°C for 45 min, followed by heat inactivation at 80 C for 15 min. An SBEreaction was performed using 1mL of purified PCR product, 0.4–2.0mM of SBE primers and a SNaPshotTM Kit (Applied Biosystems) according to the manufacturer’s instructions. Extension products were analyzed using an ABI PRISM 3130xl Genetic Analyzer and GeneMapper ID software v3.2 (Applied Biosystems). Methylation level (0–1) at each CpG site was calculated by dividing nucleotide G intensity (unconverted methylated DNA) by nucleotide G plus nucleotide A (detection of converted unmethylatedDNA) intensities.
Construction of an age-predictive model for saliva
DNA methylation profiling of saliva samples using multiplex methylation SNaPshot reaction was used for age predictive model construction. Multivariate regression analysis using the age information and DNA methylation data obtained from multiplex methylation SNaPshot reaction of the training samples was used to generate a linear predictor function composed of a set of 5 selected CpG markers to produce age as an output. Predicted ages were calculated in a set of training samples using this function. Then, to estimate the predictive accuracy of the trained model, predicted ages were also calculated using the methylation data obtained from the testing set of samples. All statistical analyses and data processing steps were performed using the statistical package (Figures 1-4).
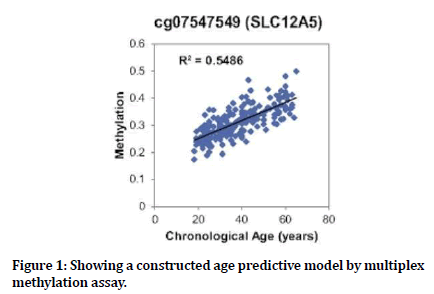
Figure 1. Showing a constructed age predictive model by multiplex methylation assay.
Figure 2. Box plot showing the association between methylation levels and CpGs, cg18384097 which is noted in gene PTPN7 showing higher methylation mean values than cg 00481951 which is noted in gene SST.
Figure 3. Box plot showing the association between methylation levels and CpGs, cg12570511 which is noted in gene TBR1 showing higher methylation mean values than cg 07547549 which is noted in gene SLC12A5.
Figure 4. Box plot showing the association between methylation levels and CpGs, cg08928145 which is noted in gene TSSK6 showing higher methylation mean values than cg 14361627 which is noted in gene KLF14.
The sensitivity of the assay was evaluated by bisulfite conversion, multiplex PCR amplification, and multiplex SBE reaction of decreasing amounts of template DNA. Saliva collected from 5 individuals aged 24, 37, 45, 52, and 64 years was selected among the samples as it represents different age groups. Genomic DNA concentration was quantified using a Quantifiler 1 Duo DNA Quantification Kit, and consecutive dilutions of DNA with concentrations ranging from 50 ng to 5 ng were subjected to bisulfite conversion using the Imprint1 DNA Modification Kit.
Bisulfite-converted DNA was eluted in 10 or 20mL of distilled water. Then, successive dilutions of bisulfite-converted DNA with concentrations ranging from 10 ng to 0.25 ng were subjected to multiplex PCR followed by multiplex SBE reaction. Peak detection to measure methylation level was performed with an analytical threshold of 50 rfu.
Results and Discussion
Selection of age-associated CpG sites from saliva
We analyzed DNA methylation profiles of saliva samples obtained from 18 to 60 years old patients using an Illumina Human Methylation 450 Bead Chip array. The number of quality-filtered CpGs was 35 and the average b-scores at these CpG sites were used to identify age-associated CpG marker candidates.
Through univariate linear regression analysis at each CpG site, a total of 35 CpGs were selected as age-associated CpG marker candidates in saliva . The selected markers showed high correlation with chronological age and a large difference in methylation between the youngest and eldest subjects. Then, to select a subset of variables (less than 10 CpGs) for use in predictive model construction, stepwise regression was implemented with data from 55 CpG sites. The stepwise regression analysis produced a model composed of 4 CpGs (cg07547549,cg14361627, cg08928145, and cg19671120 of the SLC12A5, KLF14,TSSK6, and CNGA3 genes, respectively), and all four of these CpGs were significant variables with high R2 values (0.8735, 0.8583,0.6860, and 0.7823, respectively) and a large difference between maximum and minimum b-values (>0.16) However, we decided to test more age-associated CpG marker candidates in the following targeted bisulfite sequencing analysis and selected 2 additional highly informative markers, cg00481951 and cg12757011, based on practical considerations regarding the design of bisulfite sequencing PCR primers. These 2 markers are located in the SST and TBR1genes and were 2 of the 6 genes observed more than once in the list of 35 ageassociated CpG marker candidates. In addition, to account for the heterogeneous cell type and composition of saliva, cg18384097 of the PTPN7 gene was selected for further analysis. Many factors influence the cell type and composition of saliva as similar to the factors involved in oral disorders, conservative and endodontic procedures [29-47].
DNA methylation profiling using multiplex methylation snapshot
Age associations of the 6 selected CpG markers were investigated in an independent set of 45 individuals using a developed multiplex methylation SNaPshot assay. The DNA methylation pattern at all 6 CpG markers showed a significant correlation with age. Among them, CpGs in the KLF14 and SLC12A5 genes (cg14361627 and cg07547549) showed a very strong association between methylation and age (Spearman’s rho >0.7) and could explain 63.5% and 54.9% of age variance respectively. DNA methylation of the cell type-specific marker cg18384097 varied among individuals, ranging from 0.058 to 0.889.
Evaluation of the sensitivity of the multiplex methylation snapshot assay
To test the age prediction performance of the developed multiplex methylation SNaPshot reaction for small input amounts of DNA, we selected 5 individuals aged 24, 37, 45, 52, and 64 years and compared age prediction values obtained from varying amounts of DNA. With sufficient DNA (approximately 10 ng of converted DNA obtained from bisulfite conversion of 500 ng of genomic DNA), analysis of the 5 individuals produced age prediction results of 22.7, 38.1, 50.3, 53.6, and 53.9 years, respectively . Except for the individual aged 64 years, prediction values for all individuals were within the range of 2RMSEs. Use of smaller amounts of DNA such as 10 ng of genomic DNA or 4 ng of bisulfiteconverted DNA resulted in similar prediction values to those obtained when using more DNA.
In the present study, we identified ageassociated CpG markers from saliva through epigenome-wide screening and targeted bisulfite sequencing using the methylation SNaPshot method and developed an age-predictive model composed of 6 age-associated CpG markers and a cell type-specific CpG marker. Three of these (CpGs ofthe SLC12A5, KLF14, and SST genes) were finally selected to be among the 6 age-associated markers for model construction in saliva and showed a very high correlation between methylation and age in saliva samples. This phenomenon could be due to the presence of leukocytes in saliva (leukocytes can comprise up to 80% of saliva cells) and at the same time suggests that, for some markers, methylation levels may be highly correlated between blood and saliva, while for others, the methylation levels may be more tissue-specific.
We constructed an age-predictive model for saliva using a cell type-specific CpG marker from the PTPN7 gene and 6 CpG markers, one each from the SST, CNGA3, KLF14, TSSK6, TBR1, and SLC12A5 genes, which exhibited significant age correlation in targeted bisulfite sequencing using methylation SNaPshot as well as theBeadChip array. All 6 age-associated CpG markers showed positive correlations with chronological age, while the cell-specific marker exhibited a weak negative correlation with age.
Our model composed of 7 CpGs enabled age prediction in saliva with a MAD from chronological age of 3.2 years and 95% of error within the range of 8.3 years (2RMSEs). This prediction accuracy is the highest reported so far for saliva as well as for other body fluids and tissues. When we retrained a 6 CpG model excluding the cell type-specific marker, the model’s prediction performance decreased slightly with a MAD from chronological age of 4.1 years and 95% of the error within the range of 9.7 years. In both cases, deviation from chronological age increased with advanced age, but the case without the cell type-specific marker deviated more from chronologic age in older groups. This may be due to environmental factors such as smoking, nutrition, and disease. These results are also consistent with the study results by Hewakapuge et al. [8] reporting that the advantage of the buccal-cell-signature became evident in samples of elderly donors. In addition, since the developed multiplex methylation SNaPshot worked well with more than 4 ng of bisulfite converted DNA, our model based on DNA methylation profiling using multiplex methylation SNaPshot can be easily integrated into the routine forensic laboratory workflow.
Conclusion
A model composed of 6 selected CpG sites (cg18384097 in PTPN7, cg00481951 in SST, cg14361627 in KLF14, cg08928145 in TSSK6, cg12757011 in TBR1, and cg07547549 in SLC12A5) enabled age prediction in saliva with high accuracy and is therefore likely to be useful in forensic investigations based on saliva analysis. In addition, DNA methylation profiling using the multiplex methylation SNaPshot method produced reproducible results with a small amount of DNA (4 ng of bisulfiteconvertedDNA). This multiplex system can be integrated into the routine forensic laboratory workflow after further validation tests with various casework samples. The identified ageassociated CpGmarkers can also be used in other analysis platforms such as pyrosequencing and Next Generation Sequencing through appropriate modelling.
Clinical Significance
As saliva is the second most commonly encountered fluid in the crime scenes after blood. Study focused on age identification based on the DNA extracted from human saliva. Although age extraction from salivary DNA is not reliable as DNA extracted from blood, it still can be considered as one of the diagnostic measures in forensic odontology.
Limitations
This study involved relatively a smaller number of sample sizes and the study has not focused on the correlation between the identified age model with the gender.
Future Scope
Future studies can be conducted with larger sample size and subjects having different age groups. And the correlational analysis between identified age and gender can be performed.
Author Contributions
Keerthana-Writing; Original Draft Preparation; Sindhu Ramesh-Review and Editing.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
There are no conflicts of interest.
References
- Branicki W, Brudnik U, Kupiec T, et al. (2007) Determination of phenotype associated SNPs in the MC1R gene. J Forensic Sci 2017; 52:349–354.
- Maroñas O, Phillips C, Söchtig J, et al. Development of a forensic skin colour predictive test. Forensic science Int Genetics 2014; 13:34–44.
- Walsh S, Liu F, Ballantyne KN, et al. IrisPlex: A sensitive DNA tool for accurate prediction of blue and brown eye colour in the absence of ancestry information. Forensic Sci Int Genetics 2011; 5:170–180.
- Ren F, Li C, Xi H, et al. Estimation of human age according to telomere shortening in peripheral blood leukocytes of Tibetan. Am J Forensic Med Pathol 2009; 30:252–255.
- Tsuji A, Ishiko A, Takasaki T, et al. Estimating age of humans based on telomere shortening. Forensic Sci Int 2002; 126:197–199.
- Karlsson AO, Svensson A, Marklund A, et al. Estimating human age in forensic samples by analysis of telomere repeats. Forensic Sci Int 2008; 1:569-571.
- Barrett ELB, Burke TA, Hammers M, et al. Telomere length and dynamics predict mortality in a wild longitudinal study. Mol Ecol 2013; 22:249–259.
- Hewakapuge S, van Oorschot RAH, Lewandowski P, et al. Investigation of telomere lengths measurement by quantitative real-time PCR to predict age. Legal Med 2008; 10:236–242.
- Meissner C, Bruse P, Mohamed SA, et al. The 4977 BP deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: A useful biomarker or more? Exp Gerontol 2008; 43:645–652.
- Slijepcevic P. DNA damage response, telomere maintenance and ageing in light of the integrative model. Mechanisms Ageing Develop 2008; 129:11–16.
- Meissner C, Von Wurmb N, Oehmichen M. Detection of the age-dependent 4977 bp deletion of mitochondrial DNA. Int J Legal Med 1997; 110:288-291.
- Meissner C, von Wurmb N, Schimansky B, et al. Estimation of age at death based on quantitation of the 4977-bp deletion of human mitochondrial DNA in skeletal muscle. Forensic Sci Int 1999; 105:115–124.
- Dobberstein RC, Huppertz J, von Wurmb-Schwark N, et al. Degradation of biomolecules in artificially and naturally aged teeth: Implications for age estimation based on aspartic acid racemization and DNA analysis. Forensic Sci Int 2008; 179:181–191.
- Ohtani S, Abe I, Yamamoto T. An application of D-and L-aspartic acid mixtures as standard specimens for the chronological age estimation. J Forensic Sci 2005; 50:JFS2005128-5.
- Kayser M. Forensic DNA phenotyping: Predicting human appearance from crime scene material for investigative purposes. Forensic Sci Int 2015; 18:33–48.
- Lowenson J, Clarke S. Does the chemical instability of aspartyl and asparaginyl residues in proteins contribute to erythrocyte aging? The role of protein carboxyl methylation reactions. Blood Cells 1988; 14:103–118.
- Kayser M, de Knijff P. Improving human forensics through advances in genetics, genomics and molecular biology. Nature Reviews Genetics 2011; 12:179–192.
- Kayser M, Schneider PM. DNA-based prediction of human externally visible characteristics in forensics: Motivations, scientific challenges, and ethical considerations. Forensic Sci Int 2009; 3:154–161.
- Hewakapuge S, van Oorschot R, Baindur-Hudson S. Evaluation of mtDNA mutations to predict age. Forensic Sci Int 2008; 1:561-562.
- Meissner C, Ritz-Timme S. Molecular pathology and age estimation. Forensic Sci Int 2010; 203:34–43.
- Yi SH, Xu LC, Mei K, et al. Isolation and identification of age-related DNA methylation markers for forensic age-prediction. Forensic Sci Int 2014; 11:117-125.
- Zubakov D, Liu F, van Zelm MC, et al. Estimating human age from T-cell DNA rearrangements. Current Biol 2010; 20:970–971.
- Bocklandt S, Lin W, Sehl ME, et al. Epigenetic predictor of age. PLOS One 201; 6:e14821.
- Zbieć-Piekarska R, Spólnicka M, Kupiec T, et al. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci Int 2015; 14:161–167.
- Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013; 49:359-367.
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013; 14:R115.
- Lee HY, Jung S-E, Oh YN, et al. Epigenetic age signatures in the forensically relevant body fluid of semen: a preliminary study. Forensic Sci Int 2015; 19:28–34.
- Weidner CI, Lin Q, Koch CM, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol 2014; 15: 24.
- Auswin MK, Ramesh S. Knowledge, attitude, and practice survey on the use of dental operating microscope in endodontics: A cross-sectional survey. Pharmacy Educ Res 2017.
- Mohanty S, Ramesh S, Priya VV, et al. Inhibition of purE gene using herbal compounds to treat oral diseases caused by oral pathogens–An in silico study. J Pharm Sci Res 2017; 9:1246.
- https://www.saudiendodj.com/article.asp?issn=1658-5984;year=2019;volume=9;issue=3;spage=235;epage=236;aulast=Teja
- Azeem RA, Sureshbabu NM. Clinical performance of direct versus indirect composite restorations in posterior teeth: A systematic review. J Conservative Dent 2018; 21:2–9.
- Govindaraju L, Neelakantan P and Gutmann JL. Effect of root canal irrigating solutions on the compressive strength of tricalcium silicate cements. Clin Oral Investigations 2017; 21:567–571.
- Janani K, Sandhya R. A survey on skills for cone beam computed tomography interpretation among endodontists for endodontic treatment procedure. Indian J Dent Res 2019; 30:834–838.
- Jenarthanan S, Subbarao C. Comparative evaluation of the efficacy of diclofenac sodium administered using different delivery routes in the management of endodontic pain: A randomized controlled clinical trial. J Conservative Dent 2018; 21:297–301.
- Khandelwal A, Palanivelu A. Correlation between dental caries and salivary albumin in adult population in Chennai: An In Vivo study. Brazilian Dent Sci 2019; 22:228–233.
- Malli Sureshbabu N, Selvarasu K, V JK, et al. Concentrated growth factors as an ingenious biomaterial in regeneration of bony defects after periapical surgery: A report of two cases. Case Reports Dent 2019; 2019:7046203.
- Manohar MP, Sharma S. A survey of the knowledge, attitude, and awareness about the principal choice of intracanal medicaments among the general dental practitioners and nonendodontic specialists. Indian J Dent Res 2018; 29:716–720.
- Nandakumar M, Nasim I. Comparative evaluation of grape seed and cranberry extracts in preventing enamel erosion: An optical emission spectrometric analysis. J Conservative Dent 2018; 21:516–520.
- Poorni S, Srinivasan MR, Nivedhitha MS. Probiotic strains in caries prevention: A systematic review. J Conservative Dent 2019; 22:123–128.
- Rajakeerthi R, Ms N. Natural product as the storage medium for an avulsed tooth–A systematic review. Cumhuriyet Dent J 2019; 22:249–256.
- Rajendran R, Kunjusankaran RN, Sandhya R, et al. Comparative evaluation of remineralizing potential of a paste containing bioactive glass and a topical cream containing casein phosphopeptide-Amorphous calcium phosphate: An in Vitro study. Pesquisa Br Odontopediatr Clin Integrada 2019; 19:4668.
- Ramarao S, Sathyanarayanan U. CRA Grid - A preliminary development and calibration of a paper-based objectivization of caries risk assessment in undergraduate dental education. J Conservative Dent 2019; 22:185–190.
- Siddique R, Nivedhitha MS. Effectiveness of rotary and reciprocating systems on microbial reduction: A systematic review. J Conservative Dent 2019; 22(2): 114–122.
- Siddique R, Sureshbabu NM, Somasundaram J, et al. Qualitative and quantitative analysis of precipitate formation following interaction of chlorhexidine with sodium hypochlorite, neem, and tulsi. J Conservative Dent 2019; 22:40–47.
- Siddique R, Nivedhitha MS, Jacob B. Quantitative analysis for detection of toxic elements in various irrigants, their combination (precipitate), and para-chloroaniline: An inductively coupled plasma mass spectrometry study. J Conservative Dent 2019; 22:344–350.
- Teja KV, Ramesh S, Priya V. Regulation of matrix metalloproteinase-3 gene expression in inflammation: A molecular study. J Conservative Dent 2018; 21:592–596.
Author Info
Keerthana T and Sindhu Ramesh*
Department of Conservative Dentistry and Endodontics, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, Saveetha University, Chennai, IndiaCitation: Keerthana T, Sindhu Ramesh, Age Estimation Using DNA Extracted from Human Saliva as a Diagnostic Measure in Forensic Odontology, J Res Med Dent Sci, 2021, 9 (2): 132-139.
Received: 23-Sep-2020 Accepted: 02-Jan-2021
