Review Article - (2022) Volume 10, Issue 7
Identification of Helicobacter Pylori and Their Virulence Gene (cagA and vacA) in Saliva of Iraqi Adult Patients with Gastritis Using Polymerase Chain Reaction Technique
Rawya A Mahmood* and Maha A Mahmood
*Correspondence: Rawya A Mahmood, Department of Basic Science, College of Dentistry, University of Baghdad, Iraq, Email:
Abstract
Background: Helicobacter pylori is a Gram-negative spiral-shaped bacteria that infects up to 50% of the global population. It has been identified as one of the major risk factors for acute and chronic gastritis, peptic ulcer disease, gastric cancer, and low grade gastric mucosa associated lymphoid tissue lymphomas.
Objective: To detect the presence of Helicobacter pylori in saliva of patients with gastritis by Polymerase Chain Reaction (PCR) method.
Method: Un stimulated saliva samples were collected from 60 subjects (25 males, 35 females) attending the Gastroenterology and Hepatology Hospital in Baghdad medical city, and they diagnosed by physicians according to endoscopic findings as positive H. pylori (patient group) and negative H. pylori (control group) as they detected by specialist histopathologic during the period from December 2020 to April 2021.
PCR was used to amplify 16S rRNA genes specific for H. pylori and, the positive isolates to 16S rRNA were subjected to amplify cagA and vacA genes.
Results: of the 30 saliva samples, 16 (53.33%) sample were positive to 16S rRNA genes in patients group, while only 3/30 (10.00%) saliva samples gave positive result to 16S rRNA genes in control group. In patients, all of the samples were positive for vacA gene 16/16 (100), while 5 of 16 was positive for cagA gene (31.25%), whereas in control group, 3/3 (100%) of saliva samples were positive for the two genes.
Conclusion: Helicobacter pylori can be found in the oral cavity in addition to and independently of their presence in the stomach.
Keywords
Helicobacter pylori , Gastritis, PCR, 16S rRNA, CagA gene, VacA geneIntroduction
Gastritis is a condition in which the stomach lining becomes inflamed due to mucosal injury [1]. The most prevalent infectious etiological agent linked to gastritis is Helicobacter pylori [2]. Gastritis can also be caused by long-term use of Non-Steroidal Anti-Inflammatory Medications (NSAIDs), tobacco usage, alcohol consumption and steroids [3]. Gastritis classified into two types, either acute or chronic; after H. pylori enters the stomach, it causes acute gastritis, which is defined as a temporary mild sickness marked by nausea, vomiting, epigastric discomfort, and heartburn, these symptoms begin within the first week of infection and reach their peak severity between the 9th and 12th day of illness. After that, the symptoms improve, and after two weeks of the infection, the majority of them are gone [4]. Chronic gastritis is characterized by inflammatory cell infiltration, with lymphocytes and plasma cells predominating, as well as a large number of neutrophils [5]. Helicobacter pylori are Gram-negative spiral-shaped bacteria that affects up to half of the world's population with a higher frequency in developing nations [6]. H. pylori is a fastidious microorganism because its requires a long incubation period from 5 to 7 days under micro-aerophilic conditions and required also a selective media which contain supplement and antibiotics, so the isolation of H. pylori is consider to be difficult in a proper culture media [7]. H. pylori was identified and cultured for the first time by Barry Marshall and Robin Warren [8]. The bacteria causes’ acute and chronic gastritis in all infected individuals, as well as peptic ulcer disease, gastric cancer, and low-grade gastric Mucosa Associated Lymphoid Tissue Lymphomas (MALT) [9]. H. pylori produce a wide range of virulence factors that aid in their survival in acidic environments, motility and spatial orientation in gastric mucus, and epithelial cell adhesion [10]. The main virulence genes of Helicobacter pylori are vacuolating cytotoxin a (vacA ) and cytotoxic associated antigen a (cagA ) [11]. The cagA gene is not found in all H. pylori strains, however it has been linked to clinical outcomes such as gastritis and Peptic Ulcer Disease (PUD), as well as an increased risk of gastric cancer [12,13] The vacA gene is present in all H. pylori strains, and certain of its subtypes are linked to chronic gastric mucosal inflammation and the development of PUD [14].
Invasive and non-invasive method can be used to diagnose H. pylori infection; invasive testing includes: histology, microbiological culture, rapid urease test, and polymerase chain reaction whereas non-invasive testing includes: urea breath test, stool antigen test, and serological test [15]. By using specific target genes, molecular technologies such as Polymerase Chain Reaction (PCR) have been developed for direct detection of H. pylori in clinical samples such as biopsy and saliva. It was recorded to offer better, more accurate results and time consuming [16,17]. PCR was utilized to detect H. pylori from a variety of clinical samples using genes such as: 16S rRNA, 23S rRNA, CagA , urea and ureC [18,19]. According to some researchers, the mouth cavity can serve as an extra-gastric reservoir for H. pylori and as a result, PCR technique was used to detect H. pylori in the saliva of individuals with and without gastritis [20,21]. One of the specific targets to confirm the presence of this pathogenic bacterium is the 16S rRNA, and positive amplification of H. pylori specific DNA may be measured as direct evidence of the pathogen's presence [22-24].
Literature Review
Patients and methods
Sixty subjects enrolled in this study, suffering from different complaints such as loss of appetite, weight loss, diarrohea, vomiting, dyspepsia, epigastric pain and others, have been taking appointment for Esophagogastroduodenoscopy (OGD) in Gastroenterology and Hepatology Hospital-Baghdad medical city in the period from December 2020 to April 2021. They diagnosed by physicians according to endoscopic findings as positive H. pylori (patient group) and negative H. pylori (control group) as they detected by specialist histopathologic. The endoscopic diagnosis was done with the assistance of the consultant physician at the Endoscopy Department. The patients group consist of 30 individuals (12 male and 18 female) with a mean of age (40.60) year, while the control group were 30 individuals (13 male and 17 female) with a mean of age (35.60) year. Unstimulated saliva samples (2 ml) were collected from each fasting subject from both study groups prior to endoscopy in early morning between 7 am to 8 am in sterile graduated containers by spit technique and preserved at -20 until analysis. Each frozen saliva specimen was thawed; genomic DNA was then extracted directly, using the pro mega extraction genomic DNA kit individually.
Polymerase Chain Reaction (PCR)
After extracting DNA from saliva specimens from patients and control subjects and using specific primers, the PCR amplification process was carried out. The primer sequence for amplification of the 16S rRNA gene was as follows:
16S rRNA, F (5’-TTGGAGGGCTTAGTCTCT-3’) R (5'-AAGATTGGCTCCACTTCACA-3’) with product size 310 bp [25]. Primers were used by making working solution, 10 μl from the stock solution plus 90 μl dd H2O to obtain 100 μl working solution.
PCR working solution: Preparation of the reaction mixture on ice for amplification of 16S rRNA gene was done for a 25 μl reaction volume, as in Table 1.
| Component | Volume (μl) |
|---|---|
| One Taq (NEB®) master mix | 12.5 μl |
| DNA sample | 5 μl |
| Forward primer | 1.5 μl |
| Reverse primer | 1.5 μl |
| Nuclease-free water | 4.5 μl |
| Final volume | 25 μl |
Table 1: Components of PCR reaction mixture for amplification of 16S rRNA H. pylori . º
The thermal cycle (Eppendorf, Germany) was used in conjunction with a thermal profile. The PCR process consists of thirty two cycles, each cycle involves three steps: initial denaturation at 94ºC for 5 minutes, denaturation at 94ºC for 30 sec, annealing at 55ºC for 45 sec, extension at 72ºC for 30 sec, and a final extension step at 72ºC for 7 minutes. By using agarose gel electrophoresis, the PCR products were identified by their size. The size of the PCR products was determined by comparing them to a DNA ladder (NEB®, 1000 bp DNA, England) that contains DNA fragments of known size.
Detection of h. pylori -virulence factors’ (vacA and cagA genes)
In general, all samples positive for the polymerase chain reaction were submitted for detection of vacA , cagA genes. Detection of these virulence genes was carried out by using PCR and using specific primer. The primer sequence for amplification of the vacA gene was: F (5’-ATGGAAATACAACAAACACAC-3’) R (5’CCTGAGACCGTTCCTACAGC-3’) with product size 107 bp, and primer sequence of the cagA gene was:
F(5’-AAGAAAGGCAAGAAGCAGAAAA-3’) R(5’- ACACAGAAGACAGAGCGTTATT -3’) with product size 294 bp [27]. The Primers were used by making working solution, 10 μl from the stock solution plus 90 μl dd H2O to obtain 100 μl working solution [26]. All amplification reactions were carried out in total volumes of 25 μl, as shown in (Table 2).
| Component |
Volume (μl) |
|---|---|
OneTaq (NEB®) mastermix |
12.5 μl |
DNA sample |
5 μl |
Forward Primer |
1.5 μl |
Reverse Primer |
1.5 μl |
Nuclease-Free Water |
4.5 μl |
Final Volume |
25 μl |
Table 2: Components of PCR reaction mixture for amplification of vagA and cagA genes.
The PCR thermo cycler conditions were established as thirty two cycle, each cycle involves three steps: initial denaturation at 94oC for 5 minutes, denaturation at 94ºC for 30 sec, annealing at 47ºC for 45 sec for vacA gene, while in cagA gene the annealing temperature was at 52ºC for 45 sec, extension at 72ºC for 30 sec, and a final extension step at 72ºC for 7 minutes. By using agarose gel electrophoresis, the PCR products were identified by their size. The size of the PCR products was determined by comparing them to a DNA ladder (NEB®, 1000 bp DNA, England) that contains DNA fragments of known size.
Statistical analysis: Statistical analysis was carried out using the Statistical Package for Social Science version 21 (SPSS), (Chicago, Illinois, USA).
Detection of H. pylori DNA in saliva specimens by PCR in study groups
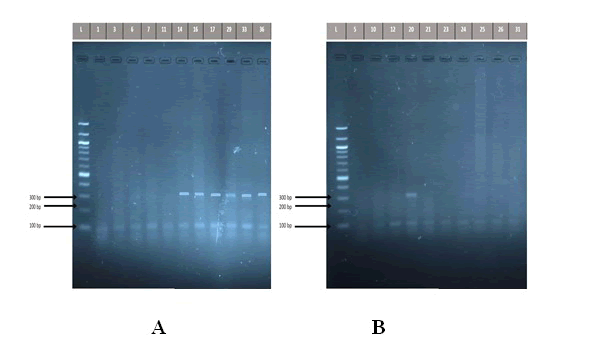
The use of saliva by some researchers in previous studies to detect the presence of H. pylori encourages us to understand the role of saliva in diagnosis of H. pylori infection as an easy and comfortable method for patients instead of endoscopy, which is painful for them. The results of this study revealed that 16 (53.33%) of saliva specimens from patients complaining of severe gastritis detected positive for 16S rRNA, while only 3(10%) of individuals in the control group detected positive for 16S rRNA, a shown in (Figure 1).
Figure 1: Detection of the PCR product DNA bands of Helicobacter pylori 16S rRNA gene (310 bp) in study groups. The amplified fragments were separated by electrophoresis on a 2% agarose gel, stained with Red Safe dye at 80 volts/cm for 1 hour. DNA ladder (100 bp step). A: patients group. Lane L DNA ladder (100 bp). Lanes 14,16,17,29,33,36 shows PCR product positive to 16S rRNA gene, while Lanes 1,3,6,7,11 shows negative to 16S rRNA gene. B: control group Lane L DNA ladder (100 bp). Lanes 20 shows PCR product positive to 16S rRNA gene, while Lanes 5,10,12, 21, 23, 24, 25, 26, 31 shows negative to 16S rRNA gene.
In a closely related study, Silva and her colleagues discovered H. pylori DNA in 16 of 30 saliva samples, suggesting that saliva samples may act as a temporary reservoir for H. pylori [28]. Whereas Nagata used nested PCR to detect H. pylori in saliva samples at percentage (4.5%) of eighty-eight subjects [29].
The present study is agree with the local study by Al Thwani and Ali in 2013 who stated that they cannot be rely on saliva samples for the detection of H. pylori infection due to the low percentage of positive results they have obtained during her research, which represented (9.4%) using PCR technique [30]. In contrast, Goud revealed a successful amplification and detection of H. pylori directly from saliva samples using 16S rRNA gene by PCR in the majority of patients with gastritis in percentage of 80%, also they showed that 30% of cases indicating H. pylori in the saliva of 50% of patients who showed negative result of H. pylori in endoscopic gastric biopsy, so this may be due to the ability of these organisms to survive in the oral cavity with low numbers for a long period without colonizing the stomach or gastric walls [31].
Some researchers found no link between H. pylori in the oral cavity and H. pylori in the stomach, thus saliva samples cannot be utilized as a primary diagnostic test for H. pylori gastric infection [32]. Myriam and her colleagues used 30 saliva samples in their research, and none of the them yielded a positive PCR result, while Ahmed and his colleagues believe that the oral cavity may be a reservoir of H. pylori infection, and that oral secretion could be an essential mode of H. pylori transmission [33,34].
Identification of vacA and cagA genes of H. pylori in saliva samples
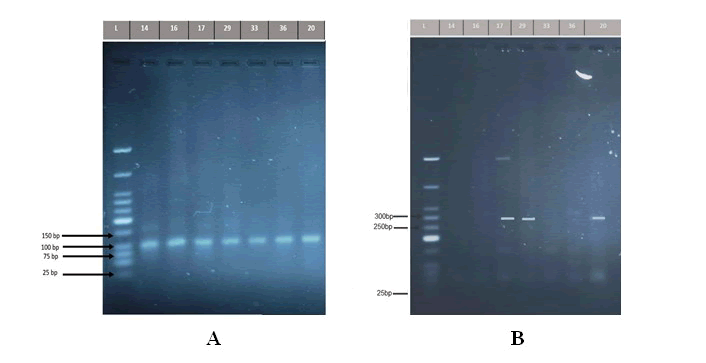
The current data revealed that the overall prevalence of the vacA gene was detected in all isolated samples of the patients 16/16 (100%), and the same results for the control group, which detected in all isolated samples 3/3 (100%) whereas the results for the cagA gene was found in only 5 saliva samples out of the 16 isolated samples of the patients, at a percentage of (31.25%), and 3/3 (100%) of cases in control group, as shown in (Figure 2).
Figure 2: A) Detection of the PCR product DNA bands of Helicobacter pylori vacA gene (107bp). The amplified fragments were separated by electrophoresis on a 2% agarose gel, stained with Red Safe dye at 80 volts/cm for 1 hour. DNA ladder (25 bp). Lane L DNA ladder (107 bp). Lanes 14,16,17,29,33,36 shows PCR product positive to vacA gene(positive H. pylori group), while Lane 20 shows PCR product positive to vacA gene (negativ H. pylori group). B) Detection of the PCR product DNA bands of Helicobacter pylori cagA gene (294 bp). The amplified fragments were separated by electrophoresis on a 2% agarose gel, stained with Red Safe dye at 80 volts/cm for 1 hour. DNA ladder (25 bp). Lane L DNA ladder (294 bp). Lanes 17,29 shows PCR product positive to cagA gene (positive H. pylori group), while Lane 20 shows PCR product positive to cagA gene (negative H. pylori group).
Results and Discussion
A comparison of the present results related to vacA and cagA gene was fluctuated with results of other studies of neighbouring countries patients, as in Jordan, Abu-Lubada and colleagues, who identified these genes from dental plaque in the oral cavity using PCR, where the cagA gene was found in 14 out of 60 samples (23.3%), while vacA gene was found in all individuals participating in this research (100%). Another research by Rasmussen and his team, who claimed that they found cagA gene in saliva sample by 13/26 (50%) and vacA gene by 26/26 (100%) and in the same year Momtaz detected the both genes at a ratio 100% in saliva samples [35-37].
Roman-Roman and colleagues discovered vacA allelic variants in 78.3% (47/60) of patients with saliva H. pylori positive, implying that gastric infection is not the only cause of H. pylori in the oral cavity, and that the bacterium could enter the mouth through other routes besides gastric reflux [38].
Miyabayashi conducted one of the earliest studies on the effect of oral H. pylori on stomach infection [39]. Their research discovered a link between gastritis caused by H. pylori infection and oral colonization of the bacteria, as well as those oral H. pylori is resistant to the standard triple anti H. pylori treatment used to remove it from the stomach. As a result, individuals with oral H. pylori had a substantially higher risk of stomach reinfection after effective treatment [40]. Many epidemiological variables, such as eating habits, oral hygiene, and illness history, were shown to influence H. pylori infection in the mouth [41]. Furthermore, the oral cavity has been proposed as a temporary or permanent H. pylori infection site.
This study showed 10% of salivary H. pylori positive cases within the individuals with gastric symptoms who showed negative status of H. pylori in histopathology biopsy may be due to production of biofilm matrix by H. pylori, which may cause embedding of H. pylori in their matrix that may would have interfered with staining of organisms leading to false negativity.
Conclusion
The presence of H. pylori in saliva samples of gastric H. pylori negative (independent, although in small numbers), and the low isolation rate (53.33%) of this bacteria from saliva of positive H. pylori gastritis patients showed a lack of connection between oral and gastric bacterial colonization. As a result, H. pylori may be detected in the oral cavity in the absence of a stomach infection.
References
- Elseweidy MM. Brief Review on the Causes, Diagnosis and Therapeutic Treatment of Gastritis Disease. Altern Integr Med 2017; 6:231.
- Jensen PJ, Feldman M, LaMont JT, et al. Acute and chronic gastritis due to Helicobacter pylori. UpToDate web site. 2019.
- Azer SA, Akhondi H. Gastritis. in Stat Pearls. Stat Pearls Publishing 2019.
- Kim N. Symptoms of Acute and Chronic H. pylori Infection. In: Kim N. (eds) Helicobacter pylori. Springer, Singapore. 2016.
- Ghasemi Basir HR, Ghobakhlou M, Akbari P, et al. Correlation between the Intensity of Helicobacter pylori colonization and severity of Gastritis. Gastroenterol Res Pract 2017; 8320496.
- Parikh NS, Ahlawat R. Helicobacter Pylori. Treasure Island (FL): Stat Pearls Publishing 2021; 30480966. [Crossref]
- Hathroubi S, Hu S, Ottemann KM. Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces. npj Biofilms Microbiomes 2020; 6:56.
- Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. The lancet 1983; 321:1273â??1275.
- Den Hoed CM, Kuipers EJ. Helicobacter pylori infection. in Hunterâ??s Tropical Medicine and Emerging Infectious Diseases. 10th edition. Science Direct 2020; 476â??480.
- Baj J, Forma A, Sitarz M, et al. Helicobacter pylori Virulence Factors Mechanisms of Bacterial Pathogenicity in the Gastric Microenvironment. Cells 2021; 10:27.
- Sedaghat H, Moniri R, Jamali R, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2, and oipA genotypes in patients with upper gastrointestinal diseases. Iran J Microbiol 2014; 6:14â??21.
- Marie MA. Relationship between Helicobacter pylori virulence genes and clinical outcomes in Saudi patients. J Korean Med Sci 2012; 27:190â??193.
- Kim SS, Ruiz VE, Carroll JD, et al. Helicobacter pylori in the pathogenesis of gastric cancer and gastric lymphoma. Cancer letters 2011; 305:228â??238.
- Siddique I, Al Qabandi A, Al Ali J, et al. Association between Helicobacter pylori genotypes and severity of chronic gastritis, peptic ulcer disease and gastric mucosal interleukin-8 levels: Evidence from a study in the Middle East. Gut pathogens 2014; 6:41.
- Ozbey G, Hanafiah A. Epidemiology, diagnosis, and risk factors of Helicobacter pylori infection in children. Euroasian J Hepatogastroenterol 2017; 7:34.
- Khalifehgholi M, Shamsipour F, Ajhdarkosh H, et al. Comparison of five diagnostic methods for Helicobacter pylori. Iran J Microbiol 2013; 5:396-401.
- Pokhrel N, Khanal B, Rai K, et al. Application of PCR and microscopy to detect Helicobacter pylori in gastric biopsy specimen among acid peptic disorders at tertiary care centre in Eastern Nepal. Can J Infect Dis Med Microbiol 2019.
- Ramis IB. Molecular methods for detection of Helicobacter pylori infection: could they be the gold standard?. J Bras Patol Med Lab 2017; 53:4.
- Jabbar HS, Adulsamed A. Gene detection of Helicobacter-pylori by use real-time PCR in patients from Wasit province: Iraq. J Entomol Zool 2018; 6:640â??643.
- Valdez Gonzalez JA, Mares-Moreno PC, Kowolik MJ, et al. Detection of Helicobacter pylori in dental plaque of Mexican children by real-time PCR. Health 2014; 6.
- Goud EVSS, Kannan R, Rao UK, et al. Identification of Helicobacter pylori in saliva of patients with and without Gastritis by Polymerase Chain Reaction. J Pharm Bioallied Sci 2019; 11:523-529.
- Hoshina S, Kahn SM, Jiang W, et al. Direct detection and amplification of Helicobacter pylori ribosomal 16S gene segments from gastric endoscopic biopsies. Diagn Microbiol Infect Dis 1990; 13:473â??479.
- Chong SKF, Lou Q, Fitzgerald JF, et al. Evaluation of 16S rRNA gene PCR with primers Hp1 and Hp2 for detection of Helicobacter pylori. J Clin Microbiol 1996; 34:2728-2730.
- Yoshida H, Hirota k, Shiratori Y, et al. Use of a gastric juice-based PCR assay todetect Helicobacter pylori infection in culture-negative patients. J Clin Microbiol 1998; 36:317â??320.
- Salman KD, Al Thwaini AN, Khalaf IA, et al. Designing of molecular tool for the detection of Helicobacterpylori in Iraqi patients using multiplex PCR technique. Ann Trop Med Public Health 2019; 22:264.
- Shetty V, Ballal M, Lingadakai R, et al. Determination of Helicobacter pylori virulence genes in clinical isolates of symptomatic patients from South Coastal Region of Karnataka A preliminary work. Austin J Gastroenterol 2015; 2:1031.
- Krashias G, Bashiardes S, Potamitou A, et al. Prevalence of Helicobacter pylori cagA and vacA genes in Cypriot patients. J Infect Dev Ctries 2013; 7:642-650.
- Silva DG, Stevens RH, Macedo JM, et al. Detection of cytotoxin genotypes of Helicobacter pylori in stomach, saliva and dental plaque. Arch Oral Biol 2009; 54:684â??688.
- Nagata R, Ohsumi T, Takenaka S, et al. Current Prevalence of Oral Helicobacter pylori among Japanese Adults Determined Using a Nested Polymerase Chain Reaction Assay. Pathogens (Basel, Switzerland) 2020; 10:10.
- Al Thwaini AN, Ali SF. Detection of helicobacter pylori in saliva and biopsy specimens of some Iraqi patients using pcr technique. International Int j adv biol 2013; 3:593-598.
- Goud EVSS, Kannan R, Rao UK, et al. Identification of Helicobacter pylori in saliva of patients with and without Gastritis by Polymerase Chain Reaction. J Pharm Bioallied Sci 2019; 11:523-529.
- Chitsazi MT, Fattahi E, Zadeh Farahani RM, et al. Helicobacter pylori in the dental plaque: is it of diagnostic value for gastric infection? Med Oral Patol Oral Cir Bucal 2006; 11:325â??328.
- Myriam A, Garza-Ramos, De La, et al. Prevalence of Helicobacer pylori in saliva and dental plaque related to periodontal disease and gastritis. Afr J Microbiol Res 2013; 7:2505-2509.
- Ahmed KS, Khan AA, Ahmed I, et al. Prevalence study to elucidate the transmission pathways of Helicobacter pylori at oral and gastroduodenal sites of a South Indian population. Singapore Med J 2006; 47:291â??296.
- Abu-Lubad M, Alzoubi H, Jarajreh D, et al. Analysis of Helicobacter pylori Genotypes Amongst Jordanians' Dental Plaque Samples. Gastroenterol Res 2018; 11:46â??51.
- Rasmussen LT, de Labio RW, Neto AC, et al. Detection of Helicobacter pylori in gastric biopsies, saliva and dental plaques of dyspeptic patients from Marilia, Sao Paulo, Brazil: presence of vacA and cagA genes. J Venom Anim Toxins incl Trop Dis 2012; 18:180-187. [Crossref]
- Momtaz H, Souod N, Dabiri H, et al. Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World J Gastroenterol 2012; 18:2105â??2111.
- Román Román A, Giono Cerezo S, Camorlinga Ponce M, et al. vacA genotypes of Helicobacter pylori in the oral cavity and stomach of patients with chronic gastritis and gastric ulcer. Enferm Infecc Microbiol Clin 2013; 31:130â??135.
- Miyabayashi H, Furihata K, Shimizu T, et al. Influence of oral Helicobacter pylori on the success of eradication therapy against gastric Helicobacter pylori. Helicobacter 2000; 5:30â??37.
- Sayed M, Ibrahim W, Abdel-bary SA, et al. Salivary PCR detection of Helicobacter pylori DNA in Egyptian patients with dyspepsia. Egypt J Med Hum Genet 2011; 12:211-216.
- Yu M, Zhang XY, Yu Q. Detection of oral Helicobacter Pylori infection using saliva test cassette. Pak J Med Sci 2015; 31:1192-1196.
- Souto R, Colombo AVP. Detection of Helicobacterpylori by polymerase chain reaction in the subgingival biofilm and saliva of nonâ?dyspeptic periodontal patients. J Periodontol 2008; 79:97â??103.
Author Info
Rawya A Mahmood* and Maha A Mahmood
Department of Basic Science, College of Dentistry, University of Baghdad, IraqCitation: Rawya A Mahmood, Maha A Mahmood, Identification of Helicobacter pylori and their virulence gene (cagA and vacA ) in Saliva of Iraqi Adult Patients with Gastritis Using Polymerase Chain Reaction Technique, J Res Med Dent Sci, 2022, 10 (7): 059-068.
Received: 28-Apr-2022, Manuscript No. JRMDS-22-P-47764; , Pre QC No. JRMDS-22-P-47764; Editor assigned: 30-Apr-2022, Pre QC No. JRMDS-22-P-47764; Reviewed: 14-May-2022, QC No. JRMDS-22-P-47764; Revised: 28-Jun-2022, Manuscript No. JRMDS-22-P-47764; Published: 05-Jul-2022