Research - (2021) Volume 9, Issue 6
Prevalence and Antimicrobial Susceptibility Pattern of Carbapenemase Producing Gram-Negative Bacterial Isolates
*Correspondence: Illamani V, Department of Microbiology, Sree Balaji Medical College and Hospital Affiliated to Bharath Institute of Higher Education and Research, India, Email:
Abstract
Carbapenems are usually the antimicrobials of choice for the treatment of MDR Gram-negative bacteria. The present study was undertaken to determine the prevalence of carbapenemase producing Gram-negative bacterial isolates from various clinical samples in Sree Balaji Medical College and Hospital, Chennai during May 2012 to August 2013. Out of the 200 samples from various wards, 96 Gram-negative bacilli were isolated from 83 (41.5%) samples. The prevalence of carbapenemase producers in this study was found to be 15.63% as detected by MHT and CDT. Among the carbapenemase producers, K. pneumonia (60%) was found to be predominant followed by E. coli (26.66%) and Pseudomonas spp. (13.33%). Out of the carbapenemase producers screened for NDM-1 by PCR, only a single isolate (6.66%) was found to possess bl a N o M- 1 gene. The rate of carbapenemase producers was found to be relatively higher in ICU. The present study informed that the emergence of antibiotics resistance.
Keywords
Antibiotic resistance, β-lactamase-producing enterobactericeae, Multi drug resistance, Hospitalized patients
Introduction
Gram-negative bacilli can cause a variety of infections 1n humans which may be either community-acquired or hospital-acquired. They include urinary tract infections (UTI), respiratory tract infections, bacteremia, septicemia, surgical site infections (SSI) and meningitis. Gramnegative bacilli have the inherent competence to produce various mechanisms which augment drug resistance and the ability to transfer the resistance determinants to other bacteria. They also can acquire resistance with the help of certain genetic materials thereby facilitating its spread [1]. The emergence of resistance in Gram-negative pathogens has alarmed many clinicians and health-care workers both in community and hospital settings. Multidrug resistant (MDR) bacteria have caused an increase in the incidence of nosocomial infections. They are often associated with prolonged and expensive treatment, which is a major drawback in developing countries [2].
The common Gram-negative bacilli causing nosocomial infections include Escherichia coli, Klebsiella species (spp.), Pseudomonas spp., Citrobacter spp., Enterobacter spp., and Acinetobacter spp. E. coli is the most common pathogen associated with UTI followed by Klebsiella spp., the latter being highly prevalent 1n respiratory tract infections such as pn eu monia. UTI, respiratory tract infections and SSI due to P. aeruginosa are the most life-threatening, particularly in the intensive care unit (ICU) [3]. The mortality rate is found to be high 1n patients with endocarditis, septicemia particularly 1n patients with malignancy, burns or drug addiction [4]. Ventilator associated pneumonia (VAP) is one of the most common nosocomial infection related to high morbidity and mortality, particularly when caused by MDR organisms and due to delayed or inappropriate antibiotic usage. Among the MDR Gram-negative pathogens, the major problem is of carbapenem resistance, where the carbapenemase production is the major defense mechanism [5]. Though there IS an Increase In frequency of antimicrobial resistance In both Gram-positive and Gram-negative pathogens, there is rapid escalation 1n resistance due to Gram-negative bacilli. New drugs such as linezolid and daptomycin are employed against infections caused by resistant Gram-positive pathogens, but treatment of infections caused by Gram-negative resistant pathogens is challenging as new drugs against them are still in the developmental stage. Further, prompt implementation of infection control is necessary to avoid dissemination of the microbes. [6,7]. Earlier, resistant Gram-negative bacterial infections were successfully treated with penicillin group of antibiotics, such as carbenicillin, ticarcillin, and piperacillin. Out of all Plactams, carbapenems have maximum antimicrobial spectrum. This is due to their high affinity for penicillin binding protein 2, good stability against most serine-based P-lactamases and excellent outer membrane permeability. Carbapenems are the antibiotics used as a last resort for treatment of MDR Gram-negative pathogens. However, resistance to this group of drugs has developed due to its increased usage [8-10].
Carbapenems currently having Food and Drug Administration (FDA) approvals for clinical use are 1m1penem, meropenem, ertapenem and doripenem. Nevertheless, bacteria can acquire carbapenemhydrolyzing P-lactamases called carbapenemases. These enzymes have emerged in various parts of the world, namely Europe, the Indian subcontinent, and the United States (US). Many of the carbapenemases recognize almost all hydrolysable P-lactams; including oxyimino Plactams and most are stable against inhibition by all Plactamase inhibitors. However, some carbapenemases are less active and they require additional mechanisms for exhibiting resistance. The commonly encountered organisms include K. pneumonia, E. coli, Pseudomonas spp., Acinetobacterspp., Citrobacter spp., and Enterobacter spp [11]. The emergence and spread of NDM-1 were reported from India, Pakistan, and UK. Later it was reported from Bangladesh, Australia [12,13]. The overall aim of this study was to obtain better information about commonly isolated MDR Gram-negative organisms from various wards of our hospital, in relation to selection and susceptibility to antimicrobial therapy. This information would help in designing antibiotic policies and would help in emergency.
Materials and Methods
The present descriptive study used the samples such as various clinical samples included 1n the study was pus, unne, endotracheal aspirates (ET aspirate), blood, sputum, ascitic fluid, high vaginal swab (HVS) and synovial fluid. The Standard reference strains such as Escherichia coli ATCC 25922 (Figure 1), Pseudomonas aeruginosa ATCC 27853, Klebsiella species (spp.) (Figure 2), and the follwing standard antibiotics including Ciprofloxacin (5μg), gentamicin (10μg), amikacin (30μg), aztreonam (30μg), cefotaxime (30μg), cefuroxime (30μg), ceftazidime (30μg), ceftriaxone (30μg), cefixime (5μg), cefdinir (5μg), piperacillin-tazobactam (100/10μg), meropenem (1Oμg) and polymyxin B (SOU) were used in the study. For isolates from urine sample- nitrofurantoin (300μg) and nalidixic acid (20μg) were included. The patients with the infected with Gram-negative bacilli (from all the routine clinical samples for diagnosis) irrespective of the ages were included in the study. The exclusion criteria as followed bacteria other than Gramnegative bacilli were excluded from this study. Permission to conduct this study was obtained from the institutional ethical committee. Various clinical samples received in microbiology laboratory during the period May 2012 to August 2013 from patients in different treating units of Sree Balaji Medical College and Hospital, Chrompet, Chennai were included in the study.
Figure 1. MacConkey agar showing colonies of Escherichia coli.
Figure 2. MacConkey agar showing colonies of Klebsiella pneumoniae.
A total of 200 clinical samples were collected. They were Gram stained and cultured on appropriate media. Out of the samples collected, a total of 96 Gram-negative bacilli identified based on the biochemical reactions (Figure 3A-G) were included in the study. Antibiotic susceptibility tests were performed. The isolates were identified by adopting the previous method88. Bio-statistical analyses were performed using SPSS 15. 0 software. Carbapenemases was detected using Modified Hodge test (MHT), Combined disc test (CDT) assays. 71, 74 The following primers were used in the PCR reactions65. NDM-Fm (5'-GGTTTGGCGATCTGGTTTTC-3'), NDM-Rm (5'-CGGAATGGCTCATCACGATC-3'), The primers amplified an internal fragment of 621 base pairs (bp) of the bl a N DM-I gene. The primers were obtained from Sigma Aldrich. PCR products after electrophoresis against a DNA ladder on gel documentation yielding a band of 621 bp were considered positive for NDM-1gene (Figure 3H).
Figure 3A. Tube catalase test.
Figure 3B. Oxidase disc test.
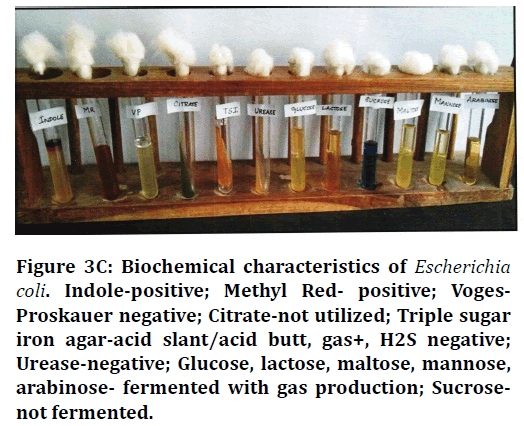
Figure 3C. Biochemical characteristics of Escherichia coli. Indole-positive; Methyl Red- positive; Voges-Proskauer negative; Citrate-not utilized; Triple sugar iron agar-acid slant/acid butt, gas+, H2S negative; Urease-negative; Glucose, lactose, maltose, mannose, arabinose- fermented with gas production; Sucrosenot fermented.
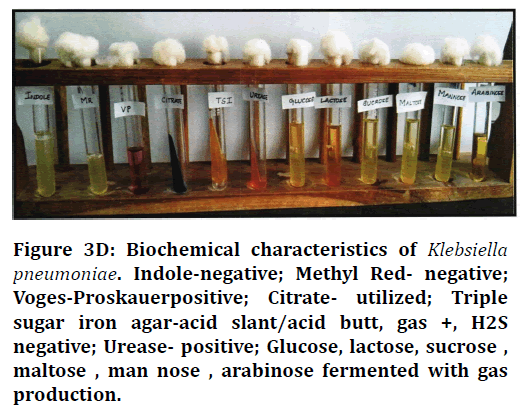
Figure 3D. Biochemical characteristics of Klebsiella pneumoniae. Indole-negative; Methyl Red- negative; Voges-Proskauerpositive; Citrate- utilized; Triple sugar iron agar-acid slant/acid butt, gas +, H2S negative; Urease- positive; Glucose, lactose, sucrose, maltose , man nose , arabinose fermented with gas production.
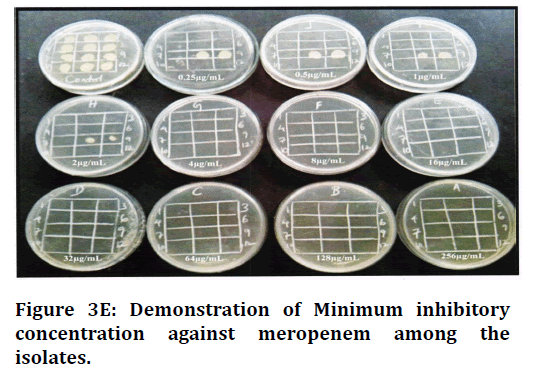
Figure 3E. Demonstration of Minimum inhibitory concentration against meropenem among the isolates.
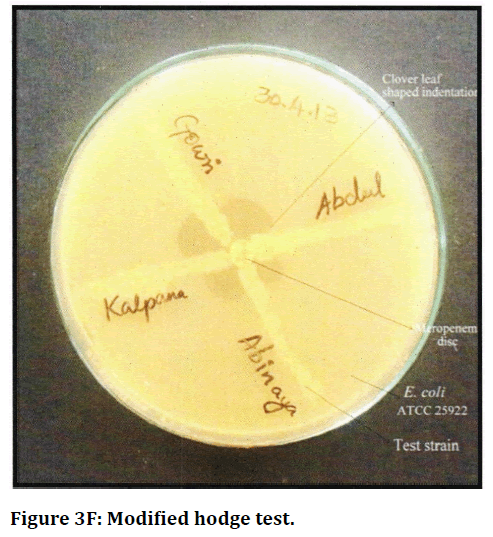
Figure 3F. Modified hodge test.
Figure 3G. Combined disc test.

Figure 3H. Gel picture of NDM-1 detection by polymerase chain reaction.
Results
Isolation of pathogens
A total of 200 clinical samples received 1n microbiology laboratory during the period May 2012 to August 2013 from patients in different treating units of our hospital were included in the study. Of 200 clinical samples, 83 (41.5%) samples yielded a total of 96 Gram negative cultures.
Of 83 samples, 70 of them showed growth of a single Gram-negative pathogen, whereas 13 samples produced 2 types of Gram-negative pathogens each. The 96 Gram negative pathogens comprised of E. coli (43.75%), Klebsiella spp. (37.5%), Pseudomonas spp. (13.54%), Proteus spp. (3.13%), and Acinetobacter spp. (2.08%). Out of 83 samples with Gram-negative bacilli growth, 43 (51.8%) were from males and 40 (48.19%) were from females (Table 1). Gram-negative pathogens (Table 2 and Figure 4) were isolated from some of the pus samples (12 out of 32) and sputum (1 out of4).
| Samples | ICU | GM | GS | Ortho | OG | Total | Samples with Gram negative bacilli growth (%) |
|---|---|---|---|---|---|---|---|
| ET aspirate | 13 | - | - | - | - | 13 | 7 (53 .84) |
| Urine | 12 | 18 | 5 | - | 13 | 48 | 29 (60.41) |
| Pus | 2 | 13 | 35 | 24 | 3 | 77 | 32 (41.55) |
| Blood | 10 | - | - | - | - | 10 | 6 (60) |
| Sputum | 1 | 6 | - | - | - | 7 | 4 (57.14) |
| Ascitic fluid | 2 | 3 | - | - | - | 5 | 2 (40) |
| HVS | - | - | - | - | 24 | 24 | 3 (12.5) |
| Synovial fluid | - | - | - | 16 | - | 16 | 0 |
| Total | 40 | 40 | 40 | 40 | 40 | 200 | 83 (41.5) |
Table 1: Sample distribution in various wards in a tertiary care hospital.
| Samples | No. of samples with Gram- negative bacilli growth | No. of Gram- negative bacilli (%) |
|---|---|---|
| ET aspirate | 7 | 7 (7.29) |
| Urine | 29 | 29 (30.20) |
| Pus | 32 | 44 (45.83) |
| Blood | 6 | 6 (6.25) |
| Sputum | 4 | 5 (5.20) |
| Ascitic fluid | 2 | 2 (2.08) |
| HVS | 3 | 3 (3.12) |
| Total | 83 | 96 |
Table 2: Details of 96 Gram-negative cultures yielded from 83 samples.
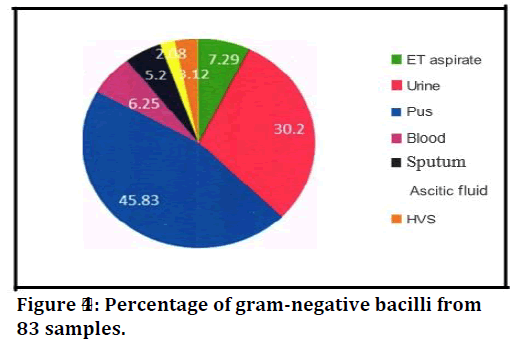
Figure 4. Percentage of gram-negative bacilli from 83 samples.
E. coli was the most prevalent organism isolated out of the total number (Table. 3) of Gram-negative bacilli followed by Klebsiella spp., Pseudomonas spp., Proteus spp. and Acinetobacter spp. Of 96 isolates, 22 (22.91%) showed resistance or intermediate susceptibility to meropenem by Kirby-Bauer disc diffusion method (Table 4).
| Organisms | No. of isolates (%) |
|---|---|
| E. coli | 42 (43.75) |
| Klebsiella spp | 36 (37.5) |
| Pseudomonas spp | 13 (13.54) |
| Proteus spp | 3 (3.125) |
| Acinetobacter spp | 2 (2 .08) |
| Total | 96 |
Table 3: Distribution, number, and percentage of 96 Gram-negative pathogens in the study.
| Organisms | No. of organisms isolated | Positive by Disc-diffusion method (%) |
|---|---|---|
| E. coli | 42 | 7 (16.67) |
| Klebsiella spp | 36 | 11 (30.55) |
| Pseudomonas spp | 13 | 4 (30.76) |
| Proteus spp | 3 | 0 |
| Cinetobacter spp | 2 | 0 |
Table 4: Result obtained in disc-diffusion method.
The MIC of meropenem was found to be: S1μg/L among 76 isolates (Susceptible) and 2μg/L among 20 isolates (Intermediate). Hence, there was no discordance between the two screening methods.
It was found that 22 isolates which showed resistance or intermediate susceptibility to meropenem by disc diffusion method were subjected to further methods namely, Modified Hodge test (MHT), and combined disc test (CDT). Of 22 isolates, 11 (50%) were positive for carbapenemase production by MHT two Klebsiella isolates, and 2 Pseudomonas isolates were found to be positive by CDT, which were not detected by MHT (Table 5 and Figure 5). It was found that 15 isolates (15.63 %) were positive for carbapenemase production by phenotypic methods (Figure 6).
| Detection of carbapenemase production | |||
|---|---|---|---|
| Meropenem resistant/intermediate susceptible isolates | MHT method (%) | CDT method (%) | Negative by both the methods (%) |
| 22 | 11 (50) | 4 (18.18) | 7 (31.81) |
Table 5: Rate of carbapenemase production among pathogens, by Modified Hodge test (MHT) and Combined disc test (CDT) phenotypic methods.
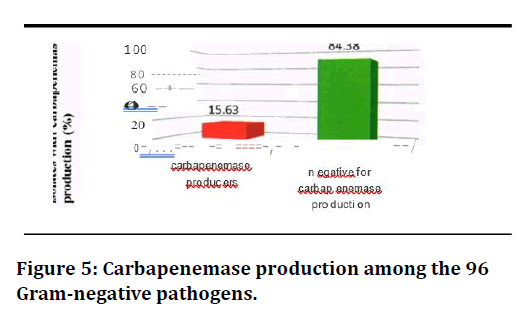
Figure 5. Carbapenemase production among the 96 Gram-negative pathogens.
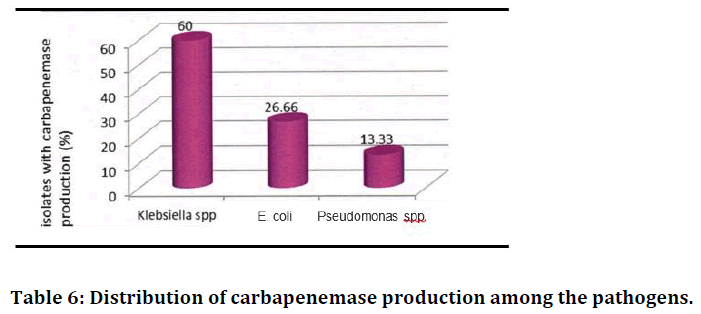
Figure 6. Distribution of carbapenemase production among the pathogens.
Overall, out of the 15 carbapenemase producers, the prevalence of carbapenemases was found to be high in Klebsiella spp. (60%), followed by E. coli (26.66%) and Pseudomonas spp. (13.33%). It was found that out of 15 carbapenemase producers, 9 (60%) and 6 (40%) were from males and females respectively; 4 (26.66%), 6 (40%), 5 (33.33%) were in the age groups of 20-40, 40-60 and above 60 (Table 6).
| Organisms | Total no. of organisms isolated | Carbapenemase producers among the pathogens (%) |
|---|---|---|
| E. coli | 42 | 4 (9.5 2%) |
| Klebsiella spp | 36 | 9 (25) |
| Pseudomonas spp | 13 | 2 (15.38) |
Table 6: Distribution of carbapenemase production among the pathogens.
Out of 36 Klebsiella isolates, nine isolates (25%) were found to produce carbapenemase. Out of 42 E. coli isolates, 4 (9.52%) and of 13 Pseudomonas isolates, two isolates (15.38%) were found to produce carbapenemase (Table 7 and Figure 7).
| Wards | No. of Gram-negative isolates | Carbapenemase producers (%) |
|---|---|---|
| ICU | 25 | 8 (32) |
| GM | 31 | 4 (12.9) |
| GS | 20 | 1 (5) |
| Ortho | 11 | 2 (18.18) |
| OG | 9 | 0 |
Table 7: Distribution of carbapenemase producers among the different wards.
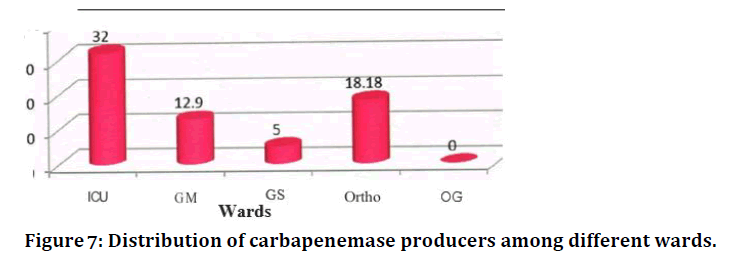
Figure 7. Distribution of carbapenemase producers among different wards.
It was found that out of the Gram negative bacilli from ICU, orthopaedics, general medicine wards and general surgery, 32 %, 18.1 8%, 12.9 % and 5% respectively were found to produce carbapenemase (Table 8 and Figure 8).
| Samples | No. of Gram-negative isolates | No. of carbapenemase producers (%) |
|---|---|---|
| ET aspirate | 7 | 3 (42.85) |
| Urine | 29 | 7 (24.13) |
| Pus | 44 | 5 (11.36) |
Table 8: Distribution of carbapenemase producers among different samples.
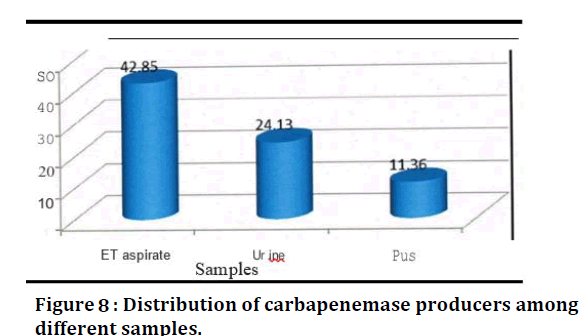
Figure 8. Distribution of carbapenemase producers among different samples.
It was found that out of the Gram-negative bacilli from ET, urine, and pus samples, 42.85 %, 24.13% and 11.36% respectively were found to produce carbapenemases (Table 9).
| Carbapenemase production | |||
|---|---|---|---|
| Samples | E. coli(%) | Klebsiella spp (%) | Pseudomonas spp (%) |
| ET | 1 (25) | 1(11.11) | 1 (50) |
| Urine | 2 (50) | 4 (44.44) | 1 (50) |
| Pus | 1 (25) | 4 (44.44) | 0 |
| Total | 4 | 9 | 2 |
Table 9: Comparison between types of samples, isolates and carbapenemase production among the pathogens.
Of fifteen patients from whom 15 carbapenemase producers were isolated, six patients (40%) were found to be associated with long term hospitalization and prolonged antibiotic usage; eight patients (53.33%) were associated with admission 1n ICU. Out of seven carbapenemase producers isolated from urine samples,two patients (28.57%) were on Foley's catheter (Table 10).
| Antibiotics | E. coli (n=4) | Klebsiella spp (n=9) | Pseudomonas spp (n=2) | |||
|---|---|---|---|---|---|---|
| R | s | R | s | R | s | |
| Ceftriaxone | 3 (7 5) | 1 (25) | 8 (88.88) | 1 (11.11) | 2 (100) | 0 |
| Ceftazidime | 4 (I 00) | 0 | 9 (100) | 0 | 1 (5 0) | 1 (5 0) |
| Cefotaxime | 2 (5 0) | 2 (50) | 5 (55.55) | 4(44.44) | 1 (5 0) | 1 (50) |
| Cefdinir | 2 (50) | 2 (50) | 4 (44.44) | 5 (55.55) | 1 (50) | 1 (5 0) |
| Cefixime | 2 (50) | 2 (50) | 5 (55.55) | 4 (44.44) | 2 (100) | 0 |
| Cefuroxime | 4 (I 00) | 0 | 7 (77.77) | 2 (22.22) | 2 (100) | 0 |
| Ciprofloxacin | 1 (2 5) | 3 (7 5) | 3 (33.33) | 6 (66.66) | 1 (5 0) | 1 (5 0) |
| Gentamicin | 2 (50) | 2 (50) | 5 (55.55) | 4 (44.44) | 1 (50) | 1 (50) |
| Amikacin | 1 (2 5) | 3 (7 5) | 2 (22.22) | 7 (77.77) | 1 (5 0) | 1 (5 0) |
| Aztreonam | 3 (7 5) | I (25) | 5 (55.55) | 4(44.44) | 0 | 2 (100) |
| Piperacillin-Tazobactam | 2 (50) | 2 (50) | 6 (66.66) | 3 (33 .33) | 2 (100) | 0 |
| Polymyxin B | 0 | 4 (100) | 0 | 9 (100) | 0 | 2 (100) |
Table 10: Antimicrobial susceptibility pattern among the meropenem resistant (carbapenemase producers).
For the carbapenemase producers isolated from unne (two E. coli, four Klebsiella spp. and one Pseudomonas spp), nalidixic acid and nitrofurantoin were tested in addition to the above-mentioned drugs. It was found that all the carbapenemase producing urine isolates except one Klebsiella spp. were resistant to nalidixic acid and nitrofurantoin (Figure 9).
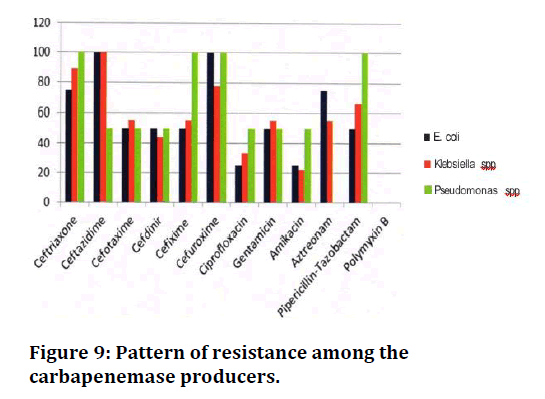
Figure 9. Pattern of resistance among the carbapenemase producers.
Detection of NDM-1 by PCR
Fifteen isolates which were detected to produce carbapenemase by phenotypic methods were screened for NDM-1 by PCR (Figure 3H). It was confirmed that a single isolate (6.66%) was found to possess bla DM-I gene. Thus, out of the carbapenemase producing K. pneumoniae, a single isolate (11.11%) was confirmed to possess bla N DM-1 gene.
Discussion
The present study was conducted to find the prevalence of carbapenemase producing Gram-negative bacilli among various clinical samples. Modified Hodge test and combined disc test which are a simple and cost effective procedure were taken as the methods of choice for phenotypic detection of carbapenemase producers.
Livermore DM et al. [8] detected that the activity of ertapenem than other carbapenems, was affected by ESBLs though it was sensitive in detection of KPC producers. According to CLSI, some Enterobacteriaceae spp., have intrinsic mechanism of resistance other than carbapenemase. Among the 96 Gram-negative bacilli, the overall prevalence of carbapenemase producers by phenotypic methods was found to be 15.63%. who reported a prevalence of 14%? A study from Chandigarh by Datta et al. [14] showed that out of 330 isolates, carbapenemase was detected 1n 26 isolates (7.87%). In previous studies it was found that out of 200 isolates, 45 (22.5%) were carbapenemase producers. In another study it is reported an overall meropenem resistance of about 30% which was higher compared to the present study.
Out of the total carbapenemase producers, the prevalence was found to be high in Klebsiella spp (60%) followed by E. coli (26.66%) and Pseudomonas spp. (13.33%). In a study it is found that Klebsiella spp. was more resistant to meropenem when compared to E. coli. This is also similar to the study from Europe who detected a higher rate of carbapenemase production in Klebsiella spp. than in E. coli.
Out of the 42 E. coli isolates from various clinical samples, carbapenemase production was found in 4 (9.52%) isolates. This is like a study from Andhra Pradesh who reported a prevalence of 11.3% among E. coli. Wattal C et al. [15] from New Delhi reported a prevalence of about 13% carbapenemase producing E. coli. A study from Morocco by M.A el Wartiti et al. [16] found a prevalence of 27. 9% carbapenemase production in Klebsiella spp. Wattal C et al. [15] from New Delhi reported a rate between 31 and 51%. But this contrasts with a study from Pondicherry by Parveen et al. [17] who documented a higher rate of 43.6% resistance to meropenem. Gupta et al. [18] from New Delhi reported a lower rate of 6.9% meropenem resistance. According to the European Antimicrobial Resistance Surveillance Network (EARS. Net, formerly EARSS) data 2009, the rates of carbapenemase producing K. pneumoniae varied from 43.5% in Greece, 17% in Cyprus, 1.3% in Italy, and 1.2% in Belgium, and less than 1 % from 23 other European countries [19]. The MHT was found to be sensitive for the detection of carbapenemases but it lacked the specificity. Also, it failed to determine the class of carbapenemases. But this test is useful for screening of carbapenemases mainly in Enterobacteriaceae with KPC, as recommended by CLSI. Therefore, in this study MHT was taken as a method of choice for screening in Entero bacteriaceae. It was found that two Klebsiella spp. which were not detected by MHT were positive by CDT. This could be due to failure of MHT in detecting weak carbapenemase producers, especially for MBL producing Enterobacteriaceae. The presence of an inhibitor such as EDTA m CDT has increased the specificity in detection of MBLs. 16 In this study, two Pseudomonas isolates (15.38%) which were positive by CDT were not detectable by MHT. This agrees with the study from Pondicherry by Noyal MIC et al. [19] who found that MHT was not preferred for determination of carbapenemase in non-fermenters and suggested the use of CDT. In previous studies it showed a carbapenemase prevalence of 15% in Pseudomonas spp. This also correlates with two other studies where the prevalence was 12% and 14% respectively. But it is in contrast to the study which documented a comparatively low prevalence of 8.05%. In the present study it was found that out of the 22 carbapenem resistant organisms by disc-diffusion method, only fifteen isolates were found to produce carbapenemase, whereas the remaining 7 (31.81%) isolates did not show a positive result by both the methods. This is in correlation with a study which showed that 7 out of 26 (26.92%) Gram-negative bacilli isolated were negative for carbapenemase production but showed resistance to carbapenem by disc-diffusion method. This could be due to other mechanisms of resistance to carbapenems such as overproduction of ESBL or Amp C -lactamase combined with porin loss. A total of 25 Gram-negative bacilli were isolated from ICU, out of which 8 (32%) carbapenemase producers were detected. Out of 31 Gram-negative isolates from general medicine ward, 4 (12.9%) were found to produce carbapenems. Among the isolates from general surgery ward, 1 out of 20 (5%) and among the isolates from orthopaedic ward, 2 out of 11 isolates (18.18%) were found to produce carbapenemase.
Recently, a study from Spain by Hidalgo L et al. [20] demonstrated that carbapenemase producers particularly NDM, were associated with increased resistance to aminoglycosides. In addition, Nordmann et al. [21] stated that many of the carbapenemase producers are frequently resistant to fluoroquinolones and aminoglycosides. Nevertheless, the rat§ of resistance to carbapenems and aminoglycosides were less compared to the other studies. In our study about 50% of the isolates exhibited resistance to gentamicin and more than 50% of the isolates were susceptible to amikacin. In one of the previous studies, it is found that resistance of 45.45% resistance to gentamicin and 40.9% resistance to amikacin which is in accordance with this study.
All the carbapenemase producers in this study (100%) were found to be susceptible to polymyxin B which agrees with the other studies. But contrasts with the studies from Uttarakhand by Thakuria et al. [22] reported 94% sensitivity to polymyxin B and from Pondicherry by Parveen et al. [17] in Klebsiella isolates who reported a very high resistance rate of approximately 60% for polymyxin B. However, in our study we did not document any resistance to polymyxin B.
In this study, it was found that only a single isolate (6.66%) was positive for blaNDM-l gene by PCR. Jamal W et al. [23] from Kuwait demonstrated that 3 (21.4%) out of 14 carbapenemase producers were found to possess blaNDM-I gene which is higher compared to the present study. According to Nagaraj et al. [24] from South India, out of 36 carbapenemase producing K. pneumoniae, 27 isolates (75%) were positive for blaNDM-l gene. But in the present study, only 11.11% of the carbapenemase producing K. pneumoniae isolates were found possess blaNDM-l gene. In a study by Bora et al. [25] from Northeastern part of India found that all the isolates of K. pneumoniae which were carbapenem resistant were found to possess blaNDM-I gene. However, none of the carbapenemase producing E. coli and Pseudomonas spp., were found to produce NDM-1 when compared to the study by Nagaraj et al. [24] who showed that 66% of E. coli were detected as NDM-1 producers. In a study by Khajuria A et al. [26] from Pune, found that out of 20 carbapenem resistant P. aeruginosa, 4 (20%) isolates were found to be positive for blaNDM-I gene which is higher compared to the present study. Thus, it implies that carbapenemase genes other than blaNDM-I gene may be the reason for carbapenemase production in the remaining isolates.
Conclusion
The study shows the prevalence of carbapenemase producers in a tertiary care hospital. The present study indicated the emergence of infections caused by carbapenemase producing pathogens and the spread of such pathogens into a hospital environment. Therefore, healthcare facilities should be vigilant in maintenance of appropriate infection control measures to decrease the current prevalence of carbapenemases.
Funding
No funding sources.
Ethical Approval
The study was approved by the Institutional Ethics Committee.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
The encouragement and support from Affiliated to Bharath Institute of Higher Education and Research, Chennai, Tamil Nadu, India is gratefully acknowledged for providing the laboratory facilities to carry out the research work.
References
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 2010; 74:417-33.
- Sharma A. Antimicrobial resistance: no action today, no cure tomorrow. Indian J Med Microbiol 2011; 29:91.
- Gaynes R, Edwards JR. National nosocomial Infections Survei11ance system. Overview of nosocomial infections caused by Gram negative bacilli. Clin Infect Dis 2005; 41:848-854.
- Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerging Infectious Diseas 2011; 17:1791.
- Dwivedi M, Mishra A, Azim A, et al. Ventilator-associated pneumonia caused by carbapenem-resistant Enterobacteriaceae carrying multiple metallo-beta-lactamase genes. Indian J Pathol Microbiol 2009; 52:339.
- Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. Infect Dis 2008; 197:1079-81.
- https://www.tandfonline.com/doi/abs/10.3109/07853890109002073
- Livermore DM. Carbapenemases. Antitimicrob Chemo Therapy 1992; 29:609-616.
- Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemotherapy 2010; 54:969-76.
- Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infectious Dis 2010; 10:597-602.
- Pitout JD. The latest threat in the war on antimicrobial resistance. Lancet Infect Dis 2010; 10:578-579.
- Centers for Disease Control and Prevention (CDC. Detection of Enterobacteriaceae isolates carrying metallo-beta-lactamase-United States, 2010. MMWR. Morbidity and mortality weekly report 2010; 59:750.
- Patel JB. Performance standards for antimicrobial susceptibility testing. Clinical and laboratory standards institute 2017.
- Datta P, Gupta V, Garg S, et al. Phenotypic method for differentiation of carbapenemases in Enterobacteriaceae: Study from north India. Indian J Pathol Microbiol 2012; 55:357.
- Wattal C, Goel N, Oberoi JK, et al. Surveillance of multidrug resistant organisms in tertiary care hospital in Delhi, India. J Assoc Physicians India 2010; 58:32-6.
- El Wartiti MA, Bahmani FZ, Elouennass M, et al. Prevalence of carbapenemase-Producing Enterobacteriaceae in a University Hospital in Rabat, Morocco: A 19-months prospective study. Int Arabic J Antimicrob Agents 2012; 2.
- Parveen RM, Harish BN, Parija SC. Emerging carbapenem resistance among nosocomial isolates of Klebsiella pneumoniae in South India. Int J Pharma Bio Sci 2010; 1.
- Gupta E, Mohanty S, Sood S, et al. Emerging resistance to carbapenems in a tertiary care hospital in north India. Indian J Med Res 2006; 124:95.
- Noyal MJ, Menezes GA, Harish BN, et al. Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative Gram-negative bacteria. Indian J Med Res 2009; 129.
- Hidalgo L, Hopkins KL, Gutierrez B, et al. Association of the novel aminoglycoside resistance determinant Rmt with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J Antimicrob Chemotherapy 2013; 68:1543-50.
- Nordmann P, Poirel L. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infection 2002; 8:321-31.
- Thakuria B, Singh P, Agrawal S, et al. Profile of infective microorganisms causing ventilator-associated pneumonia: A clinical study from resource limited intensive care unit. J Anaesthesiol Clin Pharmacol 2013; 29:361-6.
- Jamal W, Rotimi VO, Albert MJ, et al. High preva1ence of VIM-4 and NDM-I metallob-lactamases among carbapenem- resistant Enterobacteriaceae. Med Microbiol 2013; 62:1239-44.
- Nagaraj S, Chandran SP, Shamanna P, et al. Carbapenem resistance among Escherichia coli and Klebsiella pneumoniae in a tertiary care hospital in South India. Indian I Med Microb 2012; 30:93-5.
- Bora A, Ahmed G. Detection of NDM- 1 in clinical isolates of xebifea pneumontae from North -East India. J Clin Diagn Res 2012; 6:794-800.
- Khajuria A, Praharaj AK, Kumar M, et al. Emergence of NDM-1 in the clinical isolates of P. aemginosa in India. Clin Diagn Res 2013; 7:1328-133.
Author Info
1Department of Microbiology, Sree Balaji Medical College and Hospital Affiliated to Bharath Institute of Higher Education and Research, Chennai, Tamil Nadu, IndiaCitation: Aishwarya JR, Illamani V, Prevalence and Antimicrobial Susceptibility Pattern of Carbapenemase Producing Gram- Negative Bacterial Isolates, J Res Med Dent Sci, 2021, 9(6): 140-149
Received: 24-May-2021 Accepted: 16-Jun-2021
