Research - (2019) Volume 7, Issue 1
Qualitative Assessment on Isolated Tartar Forming Bacteria from Dental Caries by Pattern of Screening, Assembling and Antibiotic Sensitivity
Avra Pratim Chowdhury1*, Raunak Raj Singh2, Bhriganka Bharadwaj2, Moni P Bhuyana3 and Swarga Jyoti4
*Correspondence: Avra Pratim Chowdhury, Gauhati University, Guwahati, Assam, India, Email:
Abstract
Dental carries is a complex mixture of saliva containing bacteria which regulates oral health. Studies are subjected for determining low in caries active persons. But most investigators have been unable to confirm this finding. Here researcher wants to find out the interplay of other factors including proteins and enzymes. Thus levels and state of calcium and phosphorous in saliva may be an indicator of susceptibility to dental caries. Thus the current study is determined on the levels of salivary constituents in caries group and caries free group. As a research protocols the assembling bacterial isolation and identification assess their relationship with prevalence of dental caries.
Keywords
Anti-biogram, Dental caries, Periodontal gum, Tartar
Introduction
Oral microbiology is the study of the microorganisms of the oral cavity and their interactions between oral microorganisms or with the host. The environment present in the human mouth allows the growth of characteristic microorganisms found there. It provides a source of water and nutrients, as well as a moderate temperature. Resident microbes of the mouth adhere to the teeth and gums to resist mechanical flushing from the mouth to stomach where acid-sensitive microbes are destroyed by hydrochloric acid and food borne microbes [1,2].
The tooth surfaces are unique in that. They are the only body part not subject to metabolic turnover. Once formed, the teeth are, under the correct conditions, essentially indestructible, as witnessed by their importance in fossil records and forensic medicine. Yet in the living individual, the integrity of the teeth is assaulted by a microbial challenge so great that dental infections rank as the most universal affliction of humankind. The discomfort caused by these infections and their enormous cost (dental infections rank third in medical costs, behind heart disease and cancer, in the United States) gives dental diseases prominence despite their non-life-threatening nature [3]. Two major conditions affect teeth caries (decay) and periodontal disease [4-6].
Our tooth surface and oral cavity is good harbor of bacteria. These bacteria use the sugar (sucrose, glucose, etc.) in food to produce acids. Over time, the acids can attack the tooth, creating decay leading to a cavity and also these bacteria, along with mucus and other particles, constantly form a sticky, colorless film called “plaque” on teeth [7]. Brushing and flossing helps to get rid of plaque. Plaque when untreated with proper oral care methods can harden and form “tartar” that brushing alone doesn’t clean [8]. Only professional cleaning by a dental health professional can remove tartar [9,10].
The longer plaque and tartar are on teeth, the more harmful they become. Untreated decay can become so advanced that the tooth must be removed (extraction). The bacteria cause inflammation of the gums that is called “gingivitis” [11]. In gingivitis, the gums become red, swollen and can bleed easily. Gingivitis is a mild form of gum disease that can usually be reversed with daily brushing and flossing, and regular cleaning by a dental health professional [2,12]. This form of gum disease does not include any loss of bone and tissue that hold teeth in place [13].
Untreated gum disease can advance to “Periodontitis” (inflammation around the tooth). In periodontitis, gums pull away from the teeth and form spaces (called “pockets”) that become infected. The body’s immune system fights the bacteria as the infection spreads and grows below the gum line. Bacterial toxins and the body’s immune response to infection start to break down the bone and connective tissue that hold teeth in place. If untreated, the bones, gums, and tissue that support the teeth are degraded over time. Otherwise functional teeth may eventually become loose and have to be removed. Statistically speaking, 70% of tooth loss is due to tooth decay, 20% due to periodontal diseases and 10% due to other causes. Periodontal disease is one of the most common diseases and if untreated becomes a serious and destructive chronic infection [11,13].
However this is not the only problem. Oral health problems are also related to general health [2]. This is an important connection in terms of preventing chronic disease and disability and reducing health care costs.
Aim and Objectives of Study
The aim of the study is to ensure the pattern of antibiotic sensitivity of dental tartar forming microbes from different patients on site of periodontal gum, saliva and oral mucosa.
The objectives of the study are as follows:
• To collect the samples of dental carries from patient.
• Isolation and identification of bacteria from collected samples.
• Screening of identified microorganisms or tartar from dental caries. Comparative study and Antibiotic sensitivity test of screening samples.
Materials and Methods
Sample collection
The samples were taken mostly from the tooth region (anterior and posterior) of incisors, canine and molars of the dental patients using a sterile swab. All the patients were from the regional Regional dental college (RDC) Bhangagarh. After the samples were collected they were transferred to the respective laboratories the bacterial culture was done on the same day within 2-3 hours after the collection of the samples.
Isolation
After the samples were transferred to the respective laboratories they were dipped into peptone water for 2-5 minutes and mixed well. The samples were cultured on blood agar medium and chocolate agar medium by quadrant streaking and incubated for 24 hours at 37°C.
Liquefied agar was prepared and poured and allowed to solidify at 45°C. Swabs were dipped into peptone water and marked by the sample number as collected. 0.1 ml of bacterial suspension was transferred from the desired tubes to the sterile petri plates using sterile pipettes. Aseptically, bacterial culture was transferred in the centre of the nutrient agar plate. The inoculums were spreaded on the media using sterile glass rod. The inoculated plates were incubated at 37°C for 24 hours [14].
Isolation of bacteria
Preparation of the dilution: The microbial colonies were isolated by serial dilution plate procedure [15]. For this purpose, 5 gm of the sample was taken in 90 ml of the sterile distilled water in a sterile conical flask. It is used as the 10-1 dilution. Further dilutions were prepared up to 10-6 from the 10-1 dilution in sterile distilled water.
Media: For the isolation and enumeration of microorganisms particularly bacteria, nutrient agar media, MacConkey, Muller Hinton and CLED agar media were used although the study.
Plating: Pour plating in duplicate plates were made for each diluted sample in the following. One ml of each of the dilution was added to about 20 ml of sterile melted agar (40°C) medium in sterilized Petri plates and mixed thoroughly by rotating the plates both clockwise and anticlockwise. In this way, all plating’s were carried out from all serial diluted samples.
Incubation: After solidification of the medium, the plated were incubated (Binder Incubator) at inverted position at 37°C ± 10°C for 24 to 72 hours.
Enumeration of bacteria
After 24 hours to 48 hours of incubation, the plates having 30 to 300 colonies were selected for counting. The selected plates were placed on a colony counter (Stuart Scientific, UK) for counting is normally done by visual observation. The colonies or viable aerobic bacterial count per ml or gm were calculated by multiplying the average number of colonies per plate by the reciprocal of the dilution [16]. The calculated results would be as colony forming units (cfu) per ml or gm of sample.
Isolation of the discrete bacterial colonies
Isolation of the discrete bacterial colonies was carried out immediate after counting of the colonies. On the basis of their morphology, several different colonies were selected for the purpose. The selected colonies were marked and their characteristics were studied depending on the various points’ viz. form, elevation, margin, surface, color etc. [17,18]. Then the marked and observed bacterial colonies were transferred to nutrient agar slant aseptically for purification.
Purification of the isolates
The isolated organisms were purified through repeated plating method. Both pour plate and streak plate methods were used for the purpose. Media used for this purpose were nutrient agar (NA). When a plate yielded only one type of colony the organism were considered to be pure.
Gram's staining
A thin smear of the isolate was prepared on a glass slide, air dried and heat fixed. The heat-fixed smear was treated with aqueous crystal violet solution and was allowed to react for 1 minute. Then the slide was rinsed with tap water using wash bottle for 3-5 seconds and then treated with gram’s iodine for 1 minute and then 95% ethyl alcohol added drop by drop for 30 seconds. The smear was counter stained with safranin for 30 seconds and washed in running water. At last the slide was bottle dry and examined under immersion objective [14,19].
Biochemical test
Biochemical test were carried out according to standard protocols [14]. Biochemical tests were performed upon the gram characteristics of the colony.
Catalase test: The isolates were taken in a clean glass slide and few drops of 3% H2O2 were added to it [20].
Coagulase test
Purpose: This test is used to detect the ability of certain Staphylococcus species to clot plasma. Since coagulase production is a characteristic of the potentially pathogenic Staphylococcus aureus, it is a useful test for identifying these gram positive, catalase-positive cocci and differentiates it from the other species of Staphylococci.
Principle: Coagulase is a protein having a prothrombin like activity capable of converting fibrinogen into fibrin, which results in the formation of a visible clot in a suitable test system. Coagulase is believed to function in vivo by producing a fibrin barrier at the site of staphylococcal infection. This probably plays a role in localizing the abscesses (e.g. carbuncles and furuncles).
Coagulase is present in two forms, bound and free.
Bound coagulase (slide test): Bound coagulase, also known as clumping factor, is attached to the bacterial cell wall and is not present in the culture filtrates. Fibrin strands are formed between the bacterial cells when suspended in plasma (fibrinogen), causing them to clump into visible aggregates.
Free coagulase (tube test): Free coagulase is a thrombin-like substance present in culture filtrates. A suspension of coagulase-producing organisms is prepared in plasma in a test tube, a visible clot forms as the result of coagulase reacting with a serum substance to form a complex which, in turn, reacts with fibrinogen to produce the fibrin clot.
Media and reagents
Rabbit plasma with EDTA diluted suspension (commercially available in lyophilized form).
Procedure
Slide test
• Sufficient bacteria are emulsified in a drop of water on a microscope slide to yield a thick suspension.
• The suspension is stirred with a straight wire that has been dipped in suitable plasma.
Interpretation: A positive reaction is indicated by clumping within 5 s.
Tube test
• To two drops of an overnight broth culture in a tube (One loopful of organisms from a solid medium is also suitable) add 0.5 ml plasma diluted 1:5 with sterile physiological saline and mix.
• Bring to 37°C in the incubator or place in a 37°C water bath for 2 hours.
Interpretation: If the strain is coagulase positive, the plasma will clot usually within 4 h. Observe again at 24 h. Partial clotting is considered a positive test.
Urease production
A modified YS broth with 0.1% yeast extract and 0.0012% phenol red (w/v) was prepared in flasks and autoclaved. Urea solution was filter sterilized and then added to the above medium making 2% concentration of urea and finally the medium was dispensed into sterile test tubes. The test cultures were inoculated and incubated at 37°C for 2-4 days.
Control tubes containing the basal medium (without urea) (a) were inoculated and (b) with urea not inoculated. An increased in alkalinity indicated by a magenta red colour was regarded as the presence of urease.
Presence of urease can be detected by another method. In this method, after incubation a concentrated HCl soaked glass rod was placed at the mouth of the test tube, if ammonia is produced it reacts with HCl and produced fume of NH4Cl (Equation 1)[21].
Ammonia+Concentrated HCl soaked glass rod→Ammonium chloride (Fume) (1)
Indole test: The isolated organisms showing Gram negative rods were included on a clean test tube containing autoclaved tryptone broth. An uninoculated tube was kept as control and incubated at 35°C for 48 hours. After incubation 1 ml of Kovac’s reagent was added and the mixture was shaken well. The tube was then allowed to stand to permit the reagent to come up to the top [14,22].
MR-VP Test: MR-VP broth was prepared. The isolated organisms were inoculated on a clean test tube containing autoclaved MR-VP broth. An un-inoculated tube was kept as control and incubated at 35°C for 48 hours. After incubation few drops of methyl red reagent was added.
The VP test was performed by 12 drops of VP Reagent I and 2-3 drops of VP Reagent II culture in MR-VP broth. After addition of VP reagent the tubes were gently shaked for 30 seconds with the caps off to expose the media to oxygen and allow the reaction to complete for 15-30 minutes [14,18].
Citrate test: This was performed in Simmon’s citrate agar. Simmon’s citrate agar was prepared, poured in a clean test tube and autoclaved and allowed to solidify in a slanting position. After cooling, the isolated organisms were included on the agar slant. An un-inoculated tube was kept as control and incubated at 35°C for 48 hours [14].
Antibiotic sensitivity test
Mueller Hinton Agar was prepared and poured into petriplates and allowed to solidify. The isolated organisms have been streaked on to the solidified agar medium by sterile swabs. The selected antibiotic discs were placed on the surface on the surface of an agar plate which has already been inoculated.
The plates were incubated for 24 hours. After 24 hours the Sensitivity of the swabbed samples was determined according to the diameter of the antibiotics with respect to the bacteria as given in the EUCAST manual of zone diameter of the antibiotics and hence sensitivity and resistivity of the bacteria was determined in the Mueller Hinton Agar and the pattern of antibiotic sensitivity was determined [14,23].
Results
After collection of forty six samples from the different dental infection sites, the researchers carried out experiments on the samples by inoculating the samples in blood agar and chocolate agar respectively. After the inoculation the blood agar and the chocolate agar media containing the streaked samples were incubated at 37°C for 24 hours.
After 24 hours the petri plates were taken out and haemolysis was observed (alpha and beta haemolysis) and colony characteristics and gram stain analysis was done. The colony characteristics were done on the basis of Bergey’s Manual of Determinative Bacteriology (Figures 1-3) [24,25].
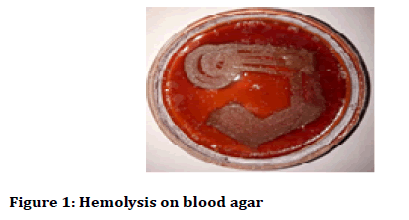
Figure 1: Hemolysis on blood agar
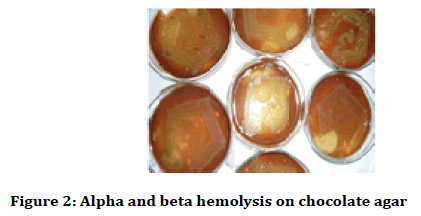
Figure 2: Alpha and beta hemolysis on chocolate agar
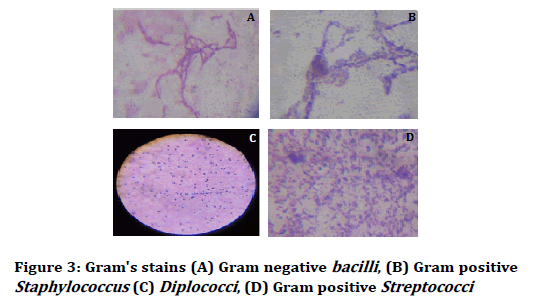
Figure 3: Gram's stains (A) Gram negative bacilli, (B) Gram positive Staphylococcus (C) Diplococci, (D) Gram positive Streptococci
The current study on “Screening and assembling of tartar forming bacteria by the pattern of antibiotic sensitivity” was performed jointly in the Biotechnology Hub, Kamrup Academy senior secondary school and in the Microbiology laboratory, Department of microbiology, Assam Down Town University. The chemicals required for the study were procured from HI media Central drug house, Bengal chemicals and Fischer Scientific. The glass wares required for the study were procured from Borosil.
Sample size: In this study the total number of samples was forty six (Table 1).
| Case No | Age (years) | Gender | Past dental problem/present illness | Diet | Antibiotic history (15 days) |
|---|---|---|---|---|---|
| 1 | 76 | M | NO | V | NO |
| 2 | 31 | M | NO | NV | NO |
| 3 | 20 | M | NO | NV | NO |
| 4 | 43 | F | Taenia solium | V | Albodangol 200 mg |
| 5 | 44 | M | Irregular bowel movement | V | NO |
| 6 | 70 | M | NO | NV | NO |
| 7 | 28 | F | NO | V | NO |
| 8 | 30 | F | NO | NV | NO |
| 9 | 20 | M | NO | NV | NO |
| 10 | 27 | M | NO | NV | Amoxilin+Clabonic acid |
| 11 | 27 | M | NO | V | NO |
| 12 | 19 | F | NO | NV | NO |
| 13 | 39 | M | NO | V | NO |
| 14 | 29 | M | NO | NV | NO |
| 15 | 85 | M | Gingival swelling | NV | NO |
| 16 | 18 | M | Recent tooth extraction | NV | NO |
| 17 | 37 | M | NO | NV | Norfloxacin |
| 18 | 22 | M | NO | NV | NO |
| 19 | 60 | F | NO | NV | NO |
| 20 | 21 | M | NO | NV | Amoxilin+Clabonic acid |
| 21 | 26 | M | NO | NV | NO |
| 22 | 36 | F | NO | NV | NO |
| 23 | 19 | F | NO | NV | NO |
| 24 | 23 | F | NO | NV | Amoxilin+Clabonic acid |
| 25 | 27 | F | NO | NV | NO |
| 26 | 30 | M | NO | NV | NO |
| 27 | 38 | F | NO | NV | NO |
| 28 | 50 | M | Dental swelling | V | Clevam |
| 29 | 22 | M | NO | NV | NO |
| 30 | 73 | M | Skin lesion+tooth extraction | V | NO |
| 31 | 50 | M | Gingival swelling+bleeding | NV | NO |
| 32 | 66 | M | Dental exaction+acidity | V | NO |
| 33 | 18 | F | NO | NV | NO |
| 34 | 52 | F | NO | NV | NO |
| 35 | 35 | M | Gingival swelling | NV | NO |
| 36 | 53 | F | Dental caries | NV | NO |
| 37 | 50 | F | NO | NV | NO |
| 38 | 30 | F | NO | NV | NO |
| 39 | 32 | M | Gingival bleeding | NV | NO |
| 40 | 24 | M | NO | NV | NO |
| 41 | 58 | M | Diabetes | NV | NO |
| 42 | 56 | F | Clinical peridontitis | V | NO |
| 43 | 50 | F | Clinical peridontitis | V | NO |
| 44 | 65 | M | Clinical peridontitis | NV | NO |
| 45 | 31 | F | Gingival bleeding | NV | NO |
| 46 | 45 | F | NO | NV | NO |
NOTE: M: Male; F: Female; V: Vegetarian; NV: Non Vegetarian; GP: Gram Positive; GN: Gram Negative
Table 1: Patient details and gram stain analysis
The ratio of samples from different patient relying on different food habituates assists the bacterial culture on regarding carbohydrate fermentative saliva and mucus.
According the tabulated studies (Table 2), most of them were cocci forming that cooperated with food particles including their enzymes. Mucilase and amylase helps to propagate the cultures as tartars in sticky form due to interact in colonized form. Decaying of dental and periodontal lime of calcium interfered by pathogenic colonized cocci and resulted in sensitivity on teeth gum and its roots. Here Table 2 was followed for further analysis comparing with patient history and with their antibiotic residues as treatment suggested by their respective Dentist.
| Gram Stain | Colony morphology |
|---|---|
| GP ROD | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GN BACILLI | Large colonies, greyish white, moist and smooth opaque |
| GP COCCI | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in chain, colonies are small circular |
| GP COCCI | Gram positive cocci arranged in chain |
| GP ROD | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP ROD | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP DIPLOCOCCI | Gram positive cocci in pairs |
| GP ROD | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GN BACCLI | Irregularly round colonies were formed along with raised, appearance |
| GP ROD | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GP ROD | Gram positive cocci arranged in cluster |
| GN BACILLI | Large colonies, greyish white, moist and smooth opaque |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP ROD | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in chain |
| GN BACILLI | Large colonies, greyish white, moist and smooth opaque |
| GP COCCI | Gram positive cocci arranged in chain |
| GP ROD | Gram positive cocci arranged in cluster |
| GN BACILLI | Mixed flora |
| GN BACILLI | Irregularly round colonies were formed along with raised, dull, appearance |
| GP ROD | Gram positive cocci arranged in cluster |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| GP COCCI | Gram positive cocci arranged in chain |
| NOTE: GP: Gram Positive; GN: Gram Negative | |
Table 2: Gram nature and colony morphology of the collected samples
On these observational studies, microscopic views were arranged accordance with morphological characteristics. The tartar forming microbes were screened in cocci form and one Gram negative (Figure 3D) in rod shape. Most of these were signified as isolated formation from food particles belongs to teeth dentin, oral mucosa, saliva and gum.
Biochemical tests (Table 3) were performed to confirm the cultural characteristics following determinative Bacteriology of Bergey's Manual [24]. So conformation of unidentified mixed growth of Pneumococcus sp., researchers applied the optosin sensitivity which was signified for resistance activities that directed inoculation of colonies on disc (Table 4). So forty six samples were resulted for resistance from serial inoculation of colonies on respect of Optosin sensitivity.
| Case no. | Indole | MR | Citrate | Catalase | Coagulase | Hemolysis | Bacitracin | Mannitol | VP |
|---|---|---|---|---|---|---|---|---|---|
| 1 | - | + | + | + | + | + | R | + | + |
| 2 | - | + | + | + | + | + | R | + | + |
| 3 | - | + | + | + | + | + | R | + | + |
| 4 | + | + | - | + | - | - | R | - | - |
| 5 | + | + | - | + | - | - | R | + | - |
| 6 | - | + | + | + | - | - | R | + | - |
| 7 | + | + | + | + | - | - | R | + | + |
| 8 | + | + | + | + | - | - | R | + | + |
| 9 | - | + | + | + | + | - | S | - | - |
| 10 | - | + | + | + | + | - | S | - | - |
| 11 | - | - | + | + | + | + | R | + | + |
| 12 | - | - | + | + | + | + | R | + | + |
| 13 | - | + | + | + | + | + | R | + | + |
| 14 | - | + | + | + | + | - | R | - | + |
| 15 | - | + | + | + | + | - | S | - | + |
| 16 | - | - | + | - | - | + | R | - | + |
| 17 | - | + | + | + | + | + | R | + | + |
| 18 | - | - | + | - | - | + | S | - | + |
| 19 | - | - | + | - | - | + | S | - | + |
| 20 | - | + | + | - | - | + | S | - | + |
| 21 | + | + | - | - | - | + | S | - | + |
| 22 | + | + | + | - | - | + | S | - | + |
| 23 | - | - | + | + | - | + | S | - | + |
| 24 | + | - | - | + | - | + | S | - | - |
| 25 | - | + | - | + | - | - | R | - | + |
| 26 | - | + | + | + | - | + | R | - | + |
| 27 | - | + | + | + | + | + | R | + | + |
| 28 | - | - | + | + | - | - | R | - | + |
| 29 | - | + | + | + | - | + | R | - | + |
| 30 | - | + | + | + | - | + | R | - | + |
| 31 | - | + | + | + | - | + | R | - | + |
| 32 | - | + | - | + | + | + | R | + | + |
| 33 | + | + | - | + | - | - | R | - | - |
| 34 | - | + | - | + | - | + | S | - | + |
| 35 | - | + | + | + | - | + | S | - | + |
| 36 | - | + | + | + | + | + | R | + | + |
| 37 | + | + | - | + | - | - | S | - | - |
| 38 | + | + | + | + | - | - | R | - | - |
| 39 | - | + | + | + | - | - | S | - | + |
| 40 | + | - | + | + | - | - | R | + | + |
| 41 | - | - | + | + | - | - | R | - | - |
| 42 | - | - | + | + | - | - | R | - | + |
| 43 | - | + | - | + | - | - | R | - | + |
| 44 | + | + | - | + | - | - | S | - | + |
| 45 | + | + | - | + | - | - | S | - | + |
| 46 | + | + | + | + | - | - | S | - | + |
NOTE: Positive (+), Negative (-), Sensitive (S), Resistant (R)
Table 3: Biochemical tests to confirm the cultural characteristics
| Case no. | Hydrolysis of urea | Optosin sensitivity |
|---|---|---|
| 1 | + | R |
| 2 | + | R |
| 3 | + | R |
| 4 | - | R |
| 5 | - | R |
| 6 | - | R |
| 7 | - | R |
| 8 | - | R |
| 9 | - | R |
| 10 | - | R |
| 11 | + | R |
| 12 | + | R |
| 13 | + | R |
| 14 | - | R |
| 15 | - | R |
| 16 | - | R |
| 17 | - | R |
| 18 | + | R |
| 19 | - | R |
| 20 | - | R |
| 21 | - | R |
| 22 | - | R |
| 23 | - | R |
| 24 | - | R |
| 25 | - | S |
| 26 | - | R |
| 27 | + | R |
| 28 | + | R |
| 29 | - | R |
| 30 | + | R |
| 31 | + | R |
| 32 | + | R |
| 33 | + | R |
| 34 | - | R |
| 35 | - | R |
| 36 | - | R |
| 37 | + | R |
| 38 | - | R |
| 39 | - | R |
| 40 | - | R |
| 41 | - | R |
| 42 | - | R |
| 43 | - | R |
| 44 | + | R |
| 45 | - | R |
| 46 | - | R |
Note: Positive (+), Negative (-), Sensitive (S), Resistant (R)
Table 4: Hydrolysis of urea and optosin sensitivity
But studying about Urea Hydrolysis, the samples 1, 2, 3, 11, 12, 13, 18, 27, 28, 30, 31, 32, 33, 37, 44 showed the positive whereas negative were selected for further analysis comparing with Pneumococcus as contaminants was present or not.
The biochemical tests were arranged by inoculating isolates from tartar particles, most of the samples were positive which changed the colour of yellow in Vogues Proskauer test (Figure 4), and not decolorized in Simmon Citrate test (Figure 5).
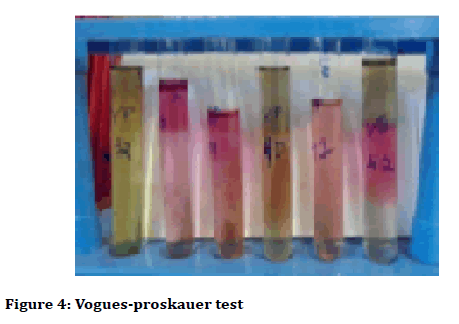
Figure 4: Vogues-proskauer test

Figure 5: Simmon citrate test
On this contrary of the studies, the outcomes of tested isolates were enlisted fruitfully for the next experiment following by the Bauer et al. [23], disc method in sensitivity determination.
The antibiotics that were set up for as sensitivity disc such as Amikacin, Chloramphenical, Ampicillin, Orfloxacin, Imipenem, Teicoplanin, Amoxiciilin and Clavulanic acid showed significant resistance against dental forming tartars (Figure 6 and Figure 7, Table 5). On the observational findings the Amikacin showed highest mark of sensitivity and Amoxycillin followed vice versa. Orfloxacin and Teicoplanin showed moderate sensitivity accordance their resistant tolerance.
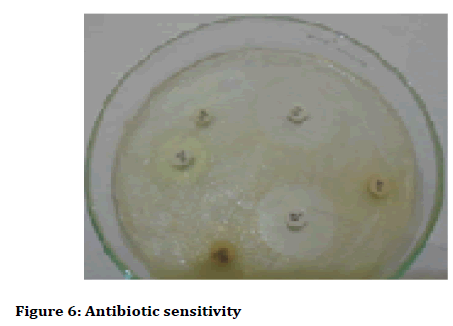
Figure 6: Antibiotic sensitivity
Figure 7: Antibiotic discs
| Case no. | AK | C | AMP | OF | IPM | TEI | AMC |
|---|---|---|---|---|---|---|---|
| 1 | S | R | R | S | R | R | R |
| 2 | R | S | R | S | R | R | R |
| 3 | S | S | R | S | R | S | R |
| 4 | S | S | R | S | R | S | R |
| 5 | R | R | R | R | R | S | R |
| 6 | S | S | R | S | S | S | R |
| 7 | S | R | R | S | R | S | R |
| 8 | S | R | R | S | R | S | R |
| 9 | S | NU | R | S | R | S | R |
| 10 | S | NU | R | S | R | R | R |
| 11 | S | NU | R | S | R | R | R |
| 12 | S | NU | R | S | NU | S | R |
| 13 | S | NU | R | S | NU | S | R |
| 14 | S | NU | R | S | NU | S | R |
| 15 | S | NU | R | S | NU | S | R |
| 16 | S | NU | R | S | R | S | R |
| 17 | S | NU | R | S | NU | S | R |
| 18 | S | NU | R | S | NU | S | R |
| 19 | S | NU | R | S | R | S | R |
| 20 | S | NU | R | S | R | R | R |
| 21 | S | NU | R | S | R | S | R |
| 22 | S | NU | R | S | R | S | R |
| 23 | S | NU | R | S | R | R | R |
| 24 | S | NU | R | S | R | S | R |
| 25 | S | NU | R | S | R | S | R |
| 26 | S | NU | R | S | NU | S | R |
| 27 | S | NU | R | S | NU | R | R |
| 28 | S | NU | R | S | NU | R | R |
| 29 | S | NU | R | S | NU | R | R |
| 30 | S | NU | R | S | NU | S | R |
| 31 | S | NU | R | S | NU | R | R |
| 32 | S | NU | R | S | R | R | R |
| 33 | S | NU | R | S | R | R | R |
| 34 | S | NU | R | S | R | R | R |
| 35 | S | NU | R | S | R | S | R |
| 36 | S | NU | R | S | R | S | R |
| 37 | S | NU | R | S | R | S | R |
| 38 | S | NU | R | S | NU | S | R |
| 39 | S | NU | R | S | R | S | R |
| 40 | S | NU | R | S | R | S | R |
| 41 | S | NU | R | S | R | S | R |
| 42 | S | NU | R | S | R | S | R |
| 43 | S | NU | R | S | R | S | R |
| 44 | S | NU | R | S | R | R | R |
| 45 | S | NU | R | S | R | R | R |
| 46 | S | NU | R | S | NU | R | R |
Note: Sensitive (S); Resistant (R); Not Used (NU)
Table 5: Antibiogram for amikacin (AK), chloramphenical (C), ampicillin (AMP), orfloxacin (OF), imipenem (IPM), teicoplanin (TEI), and amoxiciilin (AMC)
The history of patients register was strictly followed on their dentistry treatment (Table 6). Previous treatments on patients were prohibited performing this assay of Antibiotic sensitivity test.
| Case no. | Organism identified |
|---|---|
| 1 | Staphylococcus aureus |
| 2 | Staphylococcus aureus |
| 3 | Staphylococcus aureus |
| 4 | E. coli |
| 5 | Staphylococcus aureus |
| 6 | Staphylococcus aureus |
| 7 | Staphylococcus aureus |
| 8 | Staphylococcus aureus |
| 9 | Non hemolytic strepto (viridian group) |
| 10 | Non hemolytic strepto (viridian group) |
| 11 | Staphylococcus aureus |
| 12 | Staphylococcus aureus |
| 13 | Staphylococcus aureus |
| 14 | Non hemolytic strepto (viridian group) |
| 15 | Non hemolytic strepto (viridian group) |
| 16 | Streptococcus pyogenes |
| 17 | Staphylococcus aureus |
| 18 | Streptococcus pyogenes |
| 19 | Streptococcus pyogenes |
| 20 | Streptococcus pyogenes |
| 21 | Streptococcus pyogenes |
| 22 | Streptococcus pyogenes |
| 23 | Streptococcus pyogenes |
| 24 | Streptococcus pyogenes |
| 25 | Pneumococcus |
| 26 | Coagulase negative Staphylococcus (CNS) |
| 27 | Staphylococcus aureus |
| 28 | Klebsialla pneumonia |
| 29 | Coagulase negative Staphylococcus (CNS) |
| 30 | Coagulase negative Staphylococcus (CNS) |
| 31 | Coagulase negative Staphylococcus (CNS) |
| 32 | Staphylococcus aureus |
| 33 | E. coli |
| 34 | Streptococcus pyogenes |
| 35 | Streptococcus pyogenes |
| 36 | Staphylococcus aureus |
| 37 | Streptococcus pyogenes |
| 38 | E. coli |
| 39 | Streptococcus pyogenes |
| 40 | Staphylococcus aureus |
| 41 | Mixed flora |
| 42 | Klebsialla pneumonia |
| 43 | Staphylococcus aureus |
| 44 | Streptococcus pyogenes |
| 45 | Streptococcus pyogenes |
| 46 | Streptococcus pyogenes |
Table 6: Case studies and the organisms identified
Isolation and identification of pathogenic bacteria from different samples by Pour plates, Streak plate method was subjected to identify cultural characteristics in different culture media. To isolate the signified bacterial colonies in specific slant media from culture several times screening of identified bacterial count from each sample was done. Enlisted pathogenic bacteria were observed as outcomes of research and their characteristics were identified by Biochemical reactions.
Comparative study of bacterial profile was done by antibiotic sensitivity test. Following the mentioned protocols, Complying and assessment of data assembled out as result. The discussion comes out analysis of outcomes and findings. Assembling of results the research will be accomplished by methodological discussion.
Discussion
The tooth surface and oral cavity is good harbor of bacteria. These bacteria use the sugar (sucrose, glucose, etc.) in food to produce acids. Over time, the acids can attack the tooth, creating decay leading to a cavity and also these bacteria, along with mucus and other particles, constantly form a sticky, colorless film called “plaque” on teeth. Brushing and flossing helps to get rid of plaque. Plaque when untreated with proper oral care methods can harden and form “tartar” that brushing alone doesn’t clean. Only professional cleaning by a dental health professional can remove tartar. The longer plaque and tartar are on teeth, the more harmful they become.
Untreated decay can become so advanced that the tooth must be removed (extraction). The bacteria cause inflammation of the gums that is called “gingivitis”. In gingivitis, the gums become red, swollen and can bleed easily. Gingivitis is a mild form of gum disease that can usually be reversed with daily brushing and flossing, and regular cleaning by a dental health professional. This form of gum disease does not include any loss of bone and tissue that hold teeth in place.
Good dental or oral care is important to maintain healthy teeth and gums. Oral problems including bad breath, dry mouth, canker or cold sours, TMD, tooth decay, or thrush are all treatable with proper diagnosis and care. Proper nutrition means eating a well-balanced diet for good health and wellness. Low diet may contribute to periodontal disease, a major case of tooth loss in adults. When bacteria come into contact with food in mouth, acid is produced that attacks the teeth.
This can eventually lead to tooth decay, if flossing and tooth brushing are not completed on a regular basis Kuo et al. [7] studied the linkup between periodontal diseases and systemic diseases and their relationships and interactions with diabetes, respiratory diseases, cardiovascular diseases and osteoporosis in the journal Public Health Dentistry. The present study involves the screening and assembling of the tartar forming bacteria of the dental plaque of dental patients suffering from periodontitis, gum disease, pyorrhoea, and dental plaque accumulation. Tests were carried out by taking out isolates of the tartar from the anterior and posteriors mostly of incisors, canines and if present then molars also and identifying them morphologically and biochemically (Tables 7-10).
| Place of collection | Tartar sample, RDC Bhangagarh |
|---|---|
| Vegetative Cells | Cells straight rods |
| Cell Size | 1.1-1.5 × 2.0-6.0 µm |
| Spores | Spores not formed |
| Gram stain | Gram negative rod shaped |
| Acid-fast stain | Non-acid fast |
| Flagella stain | Not done |
| Motility test | Non motile |
| Agar colonies | Colonies are found in pinkish red |
| Agar slant | Mucoid growth |
| Nutrient broth | Diffused cloudiness with diffuse growth |
| Glucose broth | Flocculent growth with heavy sediment |
| Catalase activity | Positive |
| Oxidase test | Negative |
| Capsule stains | Encapsulated |
| Oxygen relation | Facultative anaerobic |
| Indole test | Positive |
| Nitrate reduction test | Positive |
| Inorganic medium | No growth |
| Citrate medium | Positive |
| Gelatin liquefaction | Not liquefied |
| Hydrogen sulphide | Negative |
| Urease test | Positive |
| Milk agar plate | Not hydrolyzed |
| Starch agar plate | Non hydrolyzed |
| Methyl red | Negative |
| V.P. test | Positive |
| Growth at | 0°C (-) |
| 10°C (+) | |
| 20°C (++) | |
| 30°C (++) | |
| 37°C (+++) | |
| 40°C (+) | |
| 45°C (+) | |
| Fermentation | Acid and Gas form: Glucose, Lactose, Sucrose, Arabinose, Mannitol and Xylose |
| No Change: Starch, Rhamnose and Raffinose | |
| Identification | Cultural, morphological and biochemical characteristics of the isolate was compared with the standard description given in Bergey’s Manual of Determinative Bacteriology [24,25] and it was found closely related with the genus Klebsiella [26]. The isolate was provisionally identified as Klebsiella pneumoni [27]. |
Table 7: Morphological, biochemical and cultural characteristics of the isolates from tartar sample (Klebsiella pneumoni)
| Place of collection | Tartar sample, RDC Bhangagarh |
|---|---|
| Vegetative Cells | Spherical or oval in cluster |
| Cell Size | 0.1-0.25 × 1-1.5 µm |
| Spores | Spores not formed |
| Gram stain | Gram positive oval shaped |
| Acid-fast stain | Non-acid fast |
| Flagella stain | Not done |
| Motility test | Non motile |
| Agar colonies | Colonies are found in yellow |
| Agar slant | Normal growth |
| Nutrient broth | Diffused cloudiness with diffuse growth |
| Glucose broth | Not used |
| Catalase activity | Positive |
| Oxidase test | Not done |
| Capsule stains | Encapsulated |
| Oxygen relation | Facultative aerobic |
| Indole test | Negative |
| Nitrate reduction test | Not done |
| Inorganic medium | No growth |
| Citrate medium | Positive |
| Gelatin liquefaction | Not liquefied |
| Hydrogen sulphide | Negative |
| Urease test | Positive |
| Milk agar plate | Not hydrolyzed |
| Starch agar plate | Not hydrolyzed |
| Methyl red | Positive |
| V.P. test | Positive |
| Growth at | 0°C (-) |
| 10°C (+) | |
| 20°C (++) | |
| 30°C (++) | |
| 37°C (+++) | |
| 40°C (+) | |
| 45°C (+) | |
| Identification | Cultural, morphological and biochemical characteristics of the isolate was compared with the standard description given in Bergey’s Manual of Determinative Bacteriology [24,25] and it was found closely related with the genus Staphylococcus [28]. The isolate was provisionally identified as Staphylococcus aureus [29]. |
Table 8: Morphological, biochemical and cultural characteristics of the isolates from tartar sample (Staphylococcus aureus)
| Place of collection | Tartar sample, RDC Bhangagarh |
|---|---|
| Vegetative Cells | Straight Cylindrical Rods |
| Cell Size | 1.1-1.5 × 2.0-6.0 µm |
| Spores | Spores not formed |
| Gram stain | Gram negative rod shaped |
| Acid-fast stain | Non-acid fast |
| Flagella stain | Not done |
| Motility test | Non motile or motile by Peritrichous flagella |
| Agar colonies | Colonies are found in pinkish red in MacConkey agar |
| Catalase activity | Positive |
| Capsule stains | Encapsulated |
| Oxygen relation | Facultative anaerobic |
| Indole test | Positive |
| Nitrate reduction test | Positive |
| Inorganic medium | No growth |
| Citrate medium | Negative |
| Gelatin liquefaction | Not liquefied |
| Hydrogen sulphide | Positive |
| Urease test | Negative |
| Methyl red | Positive |
| V.P. test | Negative |
| Growth at | 0°C (-) |
| 10°C (+) | |
| 20°C (++) | |
| 30°C (++) | |
| 37°C (+++) | |
| 40°C (+) | |
| 45°C (+) | |
| Fermentation | Acid and Gas form: Glucose, Lactose and Sucrose |
| Identification | Cultural, morphological and biochemical characteristics of the isolate was compared with the standard description given in Bergey’s Manual of Determinative Bacteriology [24,25] and it was found closely related with the genus Escherichia [30]. The isolate was provisionally identified as Escherichia coli [31]. |
Table 9: Morphological, biochemical and cultural characteristics of the isolates from tartar sample (Escherichia coli)
| Place of collection | Tartar sample, RDC Bhangagarh |
|---|---|
| Vegetative Cells | Spherical or oval |
| Cell Size | 0.5-1.0 µm (diameter) |
| Spores | Spores not formed |
| Gram stain | Gram positive chains |
| Acid-fast stain | Nonacid fast |
| Flagella stain | Not done |
| Motility test | Non motile |
| Agar colonies | Beta hemolysis |
| Catalase activity | Negative |
| Capsule stains | Encapsulated |
| Oxygen relation | Facultative anaerobic |
| Indole test | Positive |
| Inorganic medium | No growth |
| Citrate medium | Positive |
| Urease test | Negative |
| Methyl red | Positive |
| V.P. test | Positive |
| Growth at | 0°C (-) |
| 10°C (+) | |
| 20°C (++) | |
| 30°C (++) | |
| 37°C (+++) | |
| 40°C (+) | |
| 45°C (+) | |
| Identification | Cultural, morphological and biochemical characteristics of the isolate was compared with the standard description given in Bergey’s Manual of Determinative Bacteriology [24,25] and it was found closely related with the genus Streptococcus [32]. The isolate was provisionally identified as Streptococcus pyogenes [29]. |
Table 10: Morphological, biochemical and cultural characteristics of the isolates from tartar sample (Streptococcus pyogenes)
After isolation of the bacteria of the patient sample antibiotic sensitivity tests have been performed. Characterization was done by Gram’s staining and Biochemical tests.
Depending on Gram characteristics of the isolates biochemical tests such as Indole test (Figure 8), Methyl red test (Figure 9), Vogues-Proskauer test, Citrate test, Urease test and the basis of these biochemical characteristics the bacteria which was mostly found were of species Staphylococcus aureus, Escherichia coli, Nonhemolytic Streptococcus (viridian group), Streptococcus pyogenes, Pneumococcus, Coagulase negative Staphylococcus and Klebsialla pneumonia (Figure 10).

Figure 8: Indole test
Figure 9: Methyl red test
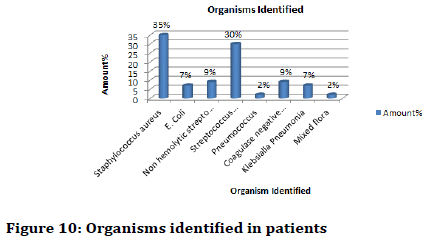
Figure 10: Organisms identified in patients
Specific antibiotic tests were performed. It was found that Staphylococcus aureus was mostly present with 35% of the patients followed by Streptococcus pyogenes contributing 30% of the patients.
Also it was found that Olfloxacin and Amkacin was the most effective antibiotic against Staphylococcus aureus and Streptococcus pyogenes. In terms of resistivity Amoxyclav (AMC) was the most resistant of the above mentioned bacterial species (Figure 11).
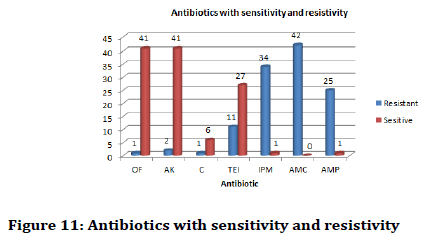
Figure 11: Antibiotics with sensitivity and resistivity
In terms of pathogenic bacteria Staphylococcus aureus was haemolytic bacteria which could cause serious disorders in other body regions (respiratory tract and gastro intestinal tract). E. coli is also pathogenic and can cause similar diseases in the gastro intestinal tract. Streptococcus pyogenesis a resident and common flora of the oral region but may cause respiratory diseases.
It was found that males had higher density of dental plaque, caries and tartar formation compared to that of females. Also it was found that the age group of 20-50 had the higher density of tartar formation along with the pathogenic bacteria in the tartar as compared to other age groups.
In terms of specific resistivity and sensitivity it was found that Staphylococcus aureus was mostly sensitive towards Olfloxacin (OF) and Amkacin (AK) and resistive towards Amoxyclav (AMC).
Hence it can be stated that patients having Staphylococcus aureus in the tartar region of the tooth are more susceptible to other infections in the human body. Also Streptococcus pyogenes which is a resident flora may become an opportunistic pathogen if bad oral health persisted in the patient. Other species like Klebsialla pneumonia was also found but the patients who were found with this species were mostly diabetic with one or two with poor oral health with excessive consumption of tobacco products. Previous studies of Iqbal et al. also stated on the linkup of diabetic patients with pathogenic bacteria in Pakistan Oral and Dental Journal which was somewhat evident in this study [4].
Overall it can be stated that more pathogenic strains of bacterial species were found in patients with poor oral health and these pathogenic stains from the oral region can cause other health problems if proper oral health care is not undertaken.
Conclusion
The present work was subjected to comparative analysis of different patient samples from reviewing doctor’s prescriptions. To collect samples as food tartars or caries in old culture from oral cavity (after having food within 12 hours), saliva and sticky mucus were most significant as isolates. Oral bacteria have evolved mechanisms to sense their environment and evade or modify the host. Bacteria occupy the ecological niche provided by both the tooth surface and gingival epithelium. So samples were screened through microbiological characteristics, biochemical properties to lay out experimentally for antibiotic sensitivity following further next step. However, a highly efficient innate host defence system constantly monitors the bacterial colonization and prevents bacterial invasion of local tissues. A dynamic equilibrium exists between dental plaque bacteria and the innate host defence system. As this aspects of research, Our research was implemented in vivo on best to review the comparative study on antibiotic sensitivity of isolated pathogenic organisms (as tartar) accordance with suggestive prescription of reputed Dentist and Doctors.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this manuscript.
References
- Aysun G, Akyüz S, Öztürk LK, et al. Salivary parameters and caries indices in children with black tooth stains. J Clin Pediatr Den 2012; 36:285-8.
- Singh S, Sharma P, Shreehari AK. Dental plaque biofilm: An invisible terror in the oral cavity. The battle against microbial pathogens: Basic science, technological advances and educational programs. Badajoz, Spain: © FORMATEX 2015; 422-8.
- Brown LJ, Oliver RC, Löe H. Periodontal diseases in the US in 1981: Prevalence, severity, extent, and role in tooth mortality. J Periodontol 1989; 60:363-70.
- Iqbal S, Kazmi F, Asad S, et al. Dental caries and diabetes mellitus. Pak Oral Dental J 2011; 31.
- Shmuly T, Zini A, Yitschaky M, et al. Can black extrinsic tooth discoloration predict a lower caries score rate in young adults? Quintessence Int 2014; 45.
- Stookey GK. The effect of saliva on dental caries. J Am Dent Assoc 2008; 139:11S-7S.
- Kuo LC, Polson AM, Kang T. Associations between periodontal diseases and systemic diseases: A review of the inter-relationships and interactions with diabetes, respiratory diseases, cardiovascular diseases and osteoporosis. Pub Health 2008; 122:417-33.
- Lazar V, Bezirtzoglou E. Microbial biofilms. In Medical significance of microbial biofilms Workshop 2008.
- National Health Strategy. Enough to make you sick: How income and environment affect health. National Health Research Strategy research paper no. 1, 1992.
- Peterson PE. Continuous improvement of oral health in the 21st century–The approach of the WHO global oral programme. The World Health Report. Geneva: World Health Organization 2003.
- Lazăr V, Chifiriuc MC, Bucur M, et al. Investigation of dental-plaque formers biofilms by optic and confocal laser scanning microscopy and microbiological tools. Rev Med Chir Soc Med Nat Iasi 2008; 112:812-20.
- Bhat S. Black tooth stain and dental caries among Udaipur school children. Int J Public Health Dent 2010; 1:13-7.
- Avila M, Ojcius DM, Yilmaz Ö. The oral microbiota: Living with a permanent guest. DNA Cell Biol 2009; 28:405-11.
- Aneja KR. Experiments in microbiology, plant pathology and biotechnology. New Age International 2007.
- Foster EM, Nelson FE, Speck ML, et al. Dairy Microbiology; St. MacMillan and Co Ltd, London 1958.
- Collins CH, Lyne PM. Microbiological methods. Butterworth & Co. (Publishers) Ltd, London 1976.
- Eklund C, Lankford CE. Laboratory manual for general microbiology. Laboratory manual for general microbiology 1967.
- Bryan JH. DNA-protein relations during microsporogenesis of Tradescantia. Chromosoma 1950; 4:369-92.
- Hucker GJ, Conn HJ. Methods of gram staining. New York Agricultural Experiment Station 1923.
- Claus GW. Understanding microbes: A laboratory textbook for microbiology. Macmillan 1989.
- Schaad NW. Laboratory guide for identification of plant pathogenic bacteria. APS Press 1988.
- Sneath PH. Endospore-forming gram-positive rods and cocci. Bergey's Manual of Systematic Bacteriology 1986; 2:1104-207.
- Bauer AW, Perry DM, Kirby WM. Single-disk antibiotic-sensitivity testing of staphylococci: An analysis of technique and results. AMA Arch Intern Med 1959; 104:208-16.
- Smith NR, Hobbs G, Buchanan RE, et al. Bergey's manual of determinative bacteriology, 8th edn. 1974; 370-3.
- Holt JG. Shorter bergey's manual of determinative bacteriology. In Shorter bergey's manual of determinative bacteriology. Williams & Wilkings 1984.
- Schröter J. Peronosporacei. Kryptogamen-Flora von Schlesien 1886; 3:228-52.
- Trevisan V. Sul micrococco della rabia e sulla possibilita di riconoscere durante il periodo d’incubazione, dall’esame del sangue della persona morsicata, se ha contratta l’infezione rabbica. Rend Ist Lombardo Accad Sci Lett Sez 1887; 20:88-105.
- Ogston A. Ueber abscesse. Arch Klin Chir 1880; 25:588-600.
- Rosenbach AJ. Mikro-organismen bei den Wund-infections-krankheiten des Menschen. JF Bergmann 1884.
- Escherich T. Die Darmbakterien des Neugeborenen und Säuglings. I, II. Fortschritte der Medizin 1885; 3:515-22.
- Migula W. Über ein neues System der Bakterien. 1895.
- Billroth T. Coccobacteria septica. Georg Reimer, Berlin 1874; 240.
Author Info
Avra Pratim Chowdhury1*, Raunak Raj Singh2, Bhriganka Bharadwaj2, Moni P Bhuyana3 and Swarga Jyoti4
1Gauhati University, Guwahati, Assam, India2Assam Down Town University (AdtU), Guwahati, Assam, India
3SRL Diagnostics, Guwahati, Assam, India
4Department of Periodontics, Regional Dental College, Guwahati, Assam, India
Citation: Avra Pratim Chowdhury, Raunak Raj Singh, Bhriganka Bharadwaj, Moni P Bhuyana, Swarga Jyoti, Qualitative assessment on isolated tartar forming bacteria from dental caries by pattern of screening, assembling and antibiotic sensitivity, J Res Med Dent Sci, 2019, 7(1): 140-157.
Received: 26-Dec-2018 Accepted: 08-Feb-2019
