Research - (2023) Volume 11, Issue 4
Virulence genes and genomic sequence of A. baumannii from different clinical samples
Anitha M1*, Umamageswari S.S.M1 and Sumathi G2
*Correspondence: Anitha M, Department of Microbiology, Vels Medical College and Hospital, Vels University, India, Email:
Abstract
125 MDR clinical isolates of Acinetobacter baumannii were tested for ESBL, carbapenemases and MBL production using standard protocol. 59 were ESBL and 79 were Carbapenemase and MBL producing Acinetobacter baumannii isolates. The genotypic expression of the virulence factors were evaluated for 15 MDRAB (Multi Drug Resistant Acinetobacter Baumannii) and 15 susceptible isolates using conventional multiplex Polymerase Chain reaction. There was statistically significant association between MDRAB and susceptible AB in respect to carbapenamase gene (p=<1.000) and MBL gene (P=0.002). The most relevant finding of this study was the identification of blaOXA 51 gene in one of the 15 susceptible isolates was isolated. This indicates the need for testing of even susceptible isolates for virulence factors. 5 MDRAB isolates were selected for genome sequencing and it was 100% confirmed as Acinetobacter baumannii at species level. This present study has brought out the importance of virulence testing for all AB isolates by genotypic methods.
Keywords
A. baumannii, Carbapenamase, ESBLs, MBLs genes
Introduction
Acinetobacter baumannii is a leading cause of nosocomial infections with increased morbidity and mortality. A. baumannii is intrinsically resistant to most antibiotics and has the ability to acquire resistance genetic determinants from the environment as well. The emerging antibiotic resistance is a serious global concern, resulting in treatment failures and increasing healthcare costs [1]. The production of β-lactamase is the most common cause of bacterial resistance to beta-lactam antibiotics. Nowadays, understanding molecular characteristics of various antibiotic resistance mechanisms along with molecular epidemiology analysis in regions with high prevalence rates of MDRAB infection plays an essential role in developing therapeutic strategies and controlling MDRAB outbreaks, both in community and hospitals. Abuse of broad-spectrum antibiotics has been demonstrated to be a major cause for the development of drug resistance of A. baumannii. At present, a number of genes responsible for drug resistance have been identified though long-term studies on the mechanisms of bacterial resistance [2].
Multidrug resistant A. baumannii (MDRAB), defined as an A. baumannii strain resistant to at least three different groups, penicillin’s and cephalosporin’s (including inhibitor combinations), fluoroquinolones, and aminoglycosides, has emerged and has been reported worldwide to significantly increase the morbidity, mortality, and cost of treatment [3]. This organism has become increasingly resistant to broad-spectrum cephalosporins due to extended-spectrum β- lactamase produced by it. Extended Spectrum Beta Lactamases (ESBLs) area class of group A beta lactamases which result in hydrolysis of first, second, and third-generation cephalosporin’s but are inhibited by beta-lactamase inhibitors like clavulanic acid [4].
Carbapenems are considered to be one of the drugs of choice for treating Acinetobacter infections. However, increased resistance to carbapenamase class of antibiotics has been reported worldwide. Results from studies have reported carbapenem resistance rate of A. baumannii as 40-75 per cent throughout India. The antibiotic resistance by ESBLs and MBLs gene production is increasing significantly in the clinical isolates of Acinetobacter spp. around the world [5]. Studies have shown that bacteria producing ESBLs or MBLs are associated with higher mortality and morbidity. Abuse of broad spectrum antibiotics has been demonstrated to be a major cause for the development of drug resistance of A. baumannii. A number of genes responsible for drug resistance have been identified through long term studies on the mechanisms of bacterial resistance. Production of β lactamase is suggested to be associated with the bacterial resistance to penicillin, cephalosporins and carbapenems, four major categories are available for the β lactamase protein encoding genes, including narrow spectrum β lactamase, ESBLs, MBLs and oxacillinase (OXA) type carbapenemases. Acinetobacter baumannii can be resistant to carbapenem by various means but, mainly it is determined by hydrolysis of antibiotics by bacterial enzymes especially carbapenem hydrolyzing β-lactamases group of enzymes like oxacillinase which has six-subtypes which correspond to class D acquired blaOXA−23-like (OXA-23, 27, and 49), chromosomal blaOXA-51-like, blaOXA-24/40-like (blaOXA-24-26, 40 and 72), blaOXA-58-lik e, blaOXA-235-like (OXA- 235 to 237) and blaOXA-143-like enzymes and Several antibiotic resistance cases due to production of acquired blaOXA have been identified worldwide. Coexistence of various antibiotic resistance mechanisms including extrusion of drugs by active efflux pumps (encoded by various Tet genes), MBLs (IMP, VIM, NDM, SPM, GIM, and SIM), ESBLs (PER, TEM, SHV, and CTX), contributes to the increase in the number of MDRAB strains [6].
Based on sequence homology, β-lactamases are clustered into four molecular classes, A, B, C, and D. All classes of β-lactamases were identified in A. baumannii. Many studies have shown that A. baumannii has natural competence to acquired exogenous DNA, and its genome has foreign DNA at high frequencies, inferring frequent horizontal gene transfer in this pathogen. Therefore, the natural competence of A. baumannii may contribute to identification of a large number of β-lactamases in this threatening pathogen. PCR products were sequenced and identified to the species level using the BLASTN tool. The selected bacterial isolate was identified by 16s rRNA gene sequence analysis using a universal primer 27F and 1492R [7-9].
Objectives
To study the genotypic pattern of virulence factors and genomic sequence of sensitive and MDRAB and correlation with age and gender distribution
Materials and Methods
Study setting and participants
This prospective study was conducted in the Department of Microbiology, in a tertiary care hospital in Kancheepuram district. During the period of six months from July 2018 to December 2019, A total of 1588 clinical samples were collected from patients admitted in various wards of the hospital, among which 153 Acinetobacter baumannii were isolated from urine, pus, sputum and miscellaneous, body fluids (excluding blood). 125 were Multi drug resistant Acinetobacter baumannii and 28 isolates were susceptible to various antibiotics [10-13].
Ethical Approval and Informed Consent
Approval was obtained from the Institutional Ethics Committee prior to the commencement of the study. Informed consent was obtained from the study participants before collecting the samples.
Bacterial identification
The samples were inoculated on Nutrient agar, 5 percent Sheep Blood agar and Mac Conkey agar and incubated overnight aerobically at both 370C and 440C. All isolates were further processed and identified by routine bacteriological and biochemical tests as per standard methodology. In order to ensure that all isolates were pure, they were cultured three times [14]. To confirm that all these isolates belonged to Acinetobacter baumannii characteristic colonies (Non Lactose-Fermenting, Glistening, Small mucoid colonies), they were subjected to Gram stain (Gram negative coccobacilli), motility (Non-motile) and standard biochemical reactions (Catalase, Oxidase, Oxidation- fermentation test, Indole production, Methyl Red(MR), Voges–Proskauer (VP), Citrate utilization, reaction in Triple Sugar Iron medium, Mannitol Motility test, Urease activity) [15,16].
Antibiotic Susceptibility of A. Baumannii
After identification by phenotypic methods, antibiotic susceptibility was performed for each isolate by the Kirby Bauer disc diffusion method on Mueller-Hinton agar using 0.5 McFarland turbidity standards and comparing zone sizes with control strain Pseudomonas aeruginosa (ATCC 27853). The antimicrobial agent was tested by using commercially available drugs by Kirby Bauer disc diffusion method [17]. The following drugs were used - Ampicillin (10 μg), Amikacin (10 μg), Ciprofloxacin (5 μg), Norfloxacin (30 μg), Levofloxacin (5 μg), Gentamicin (10 μg), Amoxyclav(30 μg), Ceftazidime (30 μg), Cefepime(30 μg), Imipenem (10 μg), Meropenem (10 μg),Piperacillin-Tazobactam(100 μg/10 μg). Antibiotic susceptibility results were interpreted by measuring the zone diameters produced and correlating them with the Clinical and Laboratory Standards Institute (CLSI) guidelines. All 125, MDR clinical isolates of Acinetobacter baumannii were tested for ESBL, carbapenemases and MBL production as per the standard protocol. About 59 were ESBL and 79 were Carbapenemase and MBL producing Acinetobacter baumannii isolates. Phenotypic detection of extended-spectrum beta-lactamases (ESBL) production by double-disc synergy test (DDST). The isolates were screened for identification of ESBL production by adopting a modified double disc synergy test. Isolates resistant to ceftazidime and/or Cefepime were tested for ESBL production by A. baumannii by disc potentiation test. A disc of Ceftazidime (30μg) and ceftazidime + clavulanic acid (30μg/10 μg) was placed 20mm apart, centreto centre on Mueller Hinton agar plate, and was incubated overnight at 37°C. A zone difference greater than or equal to 5mm around Ceftazidime and ceftazidime + clavulanic acid was interpreted as ESBL positive isolate [18].
Phenotypic detection of carbapenemases by Modified Hodge Test (MHT)
The presence of carbapenemases in A. baumannii isolates was primarily detected using Modified Hodge test according to Clinical and Laboratory Standard Institute instructions. A 0.5 McFarland standard diluted culture of Escherichia coli ATCC 25,922 (an indicator organism sensitive to carbapenems) was swabbed on the surface of Mueller-Hinton agar plates in three different directions. Meropenem disc (10 μg) (Oxoid, Basingstoke, UK) was placed at the center of each plate. The tested isolates were streaked as a thin line from the edge of the Meropenem disk to the edge of the plate. Bacterial growth was allowed for 18 h at 37° C. Indentation in the inhibition zone of E. coli or clover leaf like shaped growth of E. coli around the Meropenem disk revealed a positive MHT which indicates that this isolate is producing a Carbapenemase. Isolates resistant to imipenem and/ or Meropenem were tested for MBL production by disc potentiation test [19].
Phenotypic Identification of Mbl Production (Disc Potentiation Test)
For the identification of MBL production, all the isolates resistant to imipenem and/or Meropenem were tested by Disc Potentiation Test. Imipenem-resistant isolates were screened for the production of MBL. The double disk method was used to detect this enzyme. Colonies from overnight cultures on blood agar plates were suspended in Mueller- Hinton broth and the turbidity standardized to equal that of a bacterial concentration of 1:100 suspensions of the 0.5 McFarland standards. Then, the suspension was streaked onto Mueller-Hinton agar plates (Hi Media, Mumbai, India). A disc of Imipenem alone (10 μg) and Imipenem (10 μg) in combination with EDTA (750 μg/disc) was placed at the distance of 20 mm (Center to Center). After overnight incubation at 35°C, a ≥ 7 mm increase in the inhibition zone of diameter around Imipenem + EDTA discs, as compared to imipenem discs alone, interpreted as indicative of MBL production.
Genotypic Identification
From 125 MDRAB isolates, ESBL.MBL producing 15 MDRAB was selected. From 28 susceptible isolates, 15 Acinetobacter baumannii were selected for genotypic identification [20].
DNA Extraction
The bacterial genomic DNA was prepared by using the lysis buffer (1% Triton X-100, 10 mM Tris pH 8, 0, 1 mM EDTA) and boiling method. Briefly, colonies were picked from a MacConkey plate and inoculated into Luria- Bertani broth (Hi- Media, Mumbai, India) and incubated at 37 °C in a shaker incubator for 18 – 24 hrs. After the incubation, cell pellet was prepared from the broth. The cell pellet was washed in molecular grade water and lysis buffer was added. The bacterial suspension was heated at 95 °C for 15 min, and then pelleted by centrifugation at 12,000 ×g for 5 min. The supernatant was transferred to fresh sterile 1, 5 ml tubes, for use as tem-plates for PCR amplification [21].
Detection of extended spectrum β-lactamase ESBL &Carbapenemase and MBL encoding genes by Conventional multiplex PCR
All 30 isolates of A. baumannii were grown overnight on blood agar and genomic DNA was extracted using boiling lysis method. Conventional multiplex PCR was done for the detection of genes encoding ESBL such as blaTEM-like, blaCTX-M, and encoded for some metallo β -lactamases genes such as blaIMP-like, blaVIM, blaNDM, and blaOXA-48. The carbapenemase genes such as blaOXA-23, and blaOXA-51 were also screened by multiplex PCR and reaction conditions as described by vijayakumar et al. The amplicons were visualized in two per cent agarose gel with staining by ethidium bromide. Known positive controls for appropriate genes were used and then visualized under the gel documentation system [22].
Sequence Analysis Using Database
5 isolates were selected from 15 MDRAB, which showed the presence of virulence markers such as ESBL, MBL for sequence analysis. The bacterial isolate was identified by 16s rRNA gene sequence analysis using universal primer 27F and 1492R. The PCR product amplified with the same primer was sequenced with Sanger sequencing method. The PCR amplicons of the resistance genes were sequenced and genes were confirmed using NCBI BLASTN programme [23].
Result
Phenotypic identification
Of the total 153 samples, 125 isolates were found to be multidrug resistant AB and 28 isolates were found to be susceptible isolates. High levels of resistance was observed to first line antibiotics by Carbapenemase and Metallo- β-lactamase 79(63%), and Extended spectrum - β-lactamase 59(52%) producing isolates.
Genotypic identification
Table 1 shows Identification of genes in MDRAB producing, ESBL, carbapenamase and MBL by conventional PCR. These genes include blaOXA-51, blaOXA-23, blaOXA-48 like, blaIMP, blaVIM, blaNDM, blaTEM, blaCTX-M genes [24].
| Target enzymes | Target genes | Primer Sequences (5′→3′) | AmpliconSize (bp) |
|---|---|---|---|
| ESBL gene | blaTEM | ATGAGTATTCAACATTTCCG | 931 |
| CCAATGCTTAATCAGTGAGC | |||
| blaCTX-M | TCTTCCAGAATAAGGAATCCC | 909 | |
| CCGTTTCCGCTATTACAAAC | |||
| Carbapenemase Genes | blaOXA-51 | TCCAAATCACAGCGCTTCAAAA | 639 |
| TGAGGCTGAACAACCCATCCA | |||
| blaOXA-23 | ACTTGCTATGTGGTTGCTTCTTCTT | 797 | |
| TTCAGCTGTTTTAATGATTTCATCA | |||
| blaOXA-48 like | GCGTGGTTAAGGATGAACAC | 438 | |
| CATCAAGTTCAACCCAACCG | |||
| MBL gene | blaVIM | GTTTGGTCGCATATCGCAAC | 389 |
| AATGCGCAGCACCAGGATAG | |||
| blaIMP | GAAGGCGTTTATGTTCATAC | 587 | |
| GTACGTTTCAAGAGTGATGC | |||
| blaNDM | GCAGCTTGTCGGCCATGCGGGC | 782 | |
| GGTCGCGAAGCTGAGCACCGCAT |
Table 1: Primers sequences, target genes and amplicon sizes by conventional PCR (n=8).
Evaluation of resistant genes and virulence factors of Acinetobacter baumannii by conventional multiplex PCR method
Based on random sampling 30 isolates from different age, gender and clinical sample groups were selected for genotypic analysis, of which 15 isolates were from 125 MDRAB, 15 isolates were selected from 28 susceptible isolates for genotypic analysis. There was no statistically significant association between MDRAB and susceptible AB in respect to ESBL gene(P=0.224)). There was statistically significant association between MDRAB and susceptible AB in respect to carbapenamase gene (p=<1.000). There was statistically significant association between MDRAB and susceptible AB in respect to MBL gene(P=0.002) (Table 2). Correlation of clinical samples of MDRAB and susceptible AB with ESBL, carbapenamase, MBL genes From 30 clinical samples, six groups of genes were identified and implicated the virulence of A.baumannii. Conventional PCR was performed for 15 isolates of MDRAB. It was observed that 15(100%) blaOXA 51 and 13(87%) blaOXA 23 genes were amplified for carbapenemase genes Further, 3(20%) blaTEM and 2(13%) blaCTX-M were amplified against ESBL gene while 8(53%) blaVIM and 6(40%) blaIMP were amplified against MBL gene. Identification of these genes is an important step in elucidating mechanisms underlying antibiotic resistance in A. baumannii. Other carbapenemase genes such as blaOXA-48 like and MBL genes such as bla NDM were not detected in any of the strains [25].
| S.no | Target enzyme | Target genes | Antimicrobial susceptibility | Positive | Negative | Fishers exact test P value |
|---|---|---|---|---|---|---|
| 1 | ESBL | blaTEM | MDRAB | 3(20) | 12(80) | 0.224 |
| Susceptible | 0 | 15(100) | ||||
| 2 | ESBL | blaCTX-M | MDRAB | 2(13) | 13(87) | 0.483 |
| Susceptible | 0 | 15(100) | ||||
| 3 | Carbapenamase gene | blaOXA-51 | MDRAB | 15(100) | 0 | P=<0.001 |
| Susceptible | 1(7) | 14(93) | ||||
| 4 | Carbapenamase gene | blaOXA-23 | MDRAB | 13 | 2(13) | P=<0.001 |
| Susceptible | 0 | 15(100) | ||||
| 5 | Carbapenamase gene | blaOXA-48 | MDRAB | 0 | 15(100) | P=1.0 |
| Susceptible | 0 | 15(100) | ||||
| 6 | MBL gene | blaVIM | MDRAB | 8(53) | 7(47) | P=0.002 |
| Susceptible | 0 | 15(100) | ||||
| 7 | MBL gene | blaIMP | MDRAB | 6(40) | 9(60) | P=0.017 |
| Susceptible | 0 | 15(100) | ||||
| 8 | MBL gene | blaNDM | MDRAB | 0 | 15(100) | P=1.0 |
| Susceptible | 0 | 15(100) |
Table 2: Identification of genes in MDRAB and susceptible AB producing ESBL, MBL by conventional PCR(n=30).
15 susceptible isolates of Acinetobacter baumannii were tested for the presence of enzyme producing genes. One of the most important features of this study was the identification of blaOXA 51 genes in one susceptible isolate and remaining blaOXA-23, blaOXA-48 like, blaIMP, blaVIM, blaNDM, blaTEM, blaCTX-M genes were not amplified in susceptible isolates. This indicates the need for testing of even susceptible isolates for virulence factors (Table 3, Figures 1 & 2).
| MDRAB(n=15) | |||||||||||||
| Clinical samples | ESBL gene | Carbapenamase gene | MBL gene | ||||||||||
| blaTEM | blaCTX-M | blaOXA-51 | blaOXA-23 | blaOXA-48 | blaVIM | blaIMP | blaNDM | ||||||
| Urine | 2 | 1 | 5 | 5 | Nil | 3 | 3 | Nil | |||||
| Pus | 1 | 1 | 5 | 4 | Nil | 3 | 2 | Nil | |||||
| Sputum | Nil | Nil | 3 | 3 | Nil | 1 | 1 | Nil | |||||
| Miscellaneous | Nil | Nil | 2 | 1 | Nil | 1 | Nil | Nil | |||||
| Total | 3 | 2 | 15 | 13 | Nil | 8 | 6 | Nil | |||||
| Susceptible AB(n=15) | |||||||||||||
| Clinical samples | ESBL gene | Carbapenamase gene | MBL gene | ||||||||||
| blaTEM | blaCTX-M | blaOXA-51 | blaOXA-23 | blaOXA-48 | blaVIM | blaIMP | blaNDM | ||||||
| Nil | Nil | Nil | Nil | Nil | Nil | Nil | Nil | ||||||
| Nil | Nil | 1 | Nil | Nil | Nil | Nil | Nil | ||||||
| Sputum | Nil | Nil | Nil | Nil | Nil | Nil | Nil | Nil | |||||
| Miscellaneous | Nil | Nil | Nil | Nil | Nil | Nil | Nil | Nil | |||||
| Total | Nil | Nil | 1 | Nil | Nil | Nil | Nil | Nil | |||||
Table 3: Correlation of clinical samples of MDRAB and susceptible AB with ESBL, carbapenamase, MBL genes (n=30).
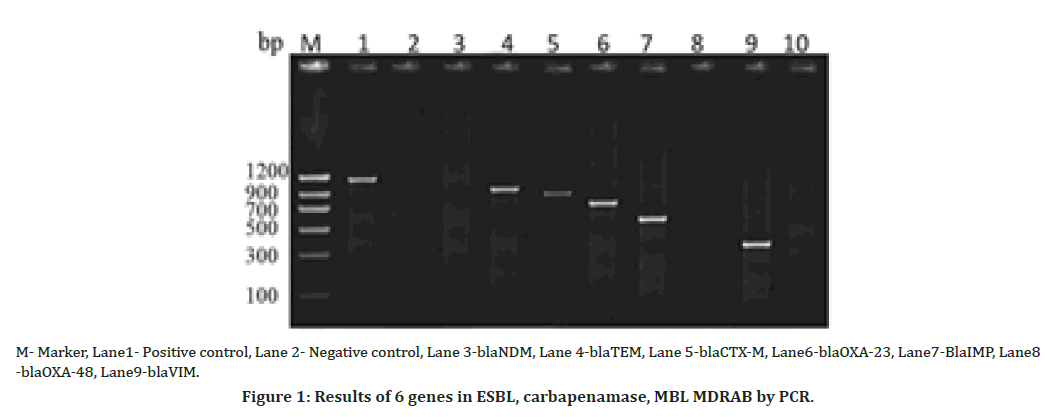
Figure 1. Results of 6 genes in ESBL, carbapenamase, MBL MDRAB by PCR.
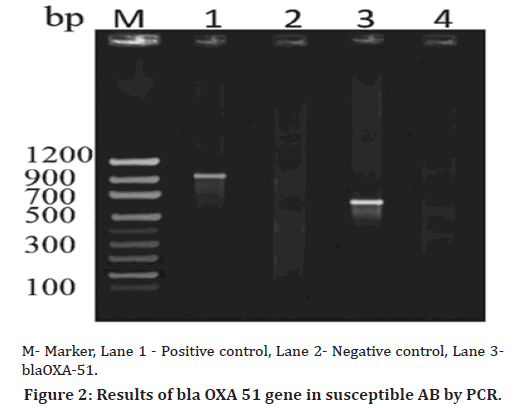
Figure 2. Results of bla OXA 51 gene in susceptible AB by PCR.
Genome Sequence Analysis
The PCR product was amplified using universal primer 27F and 1492R. Sequencing was done with Sanger sequencing method (Eurofins India Ltd., Bangalore) and obtained sequence was identified using a BLASTN tool from NCBI. 5 isolates from 15 MDRAB, isolates were randomly selected for sequence analysis from samples of urine, pus and sputum. All 5 isolates were positive for genome sequence analysis of Acinetobacter baumannii.
Among 15 susceptible isolates, one isolate from pus sample from a female patient was positive for blaoxa 51 gene. The obtained sequence was submitted to NCBI and accession number was obtained (Accession No: MW582860) [26]. One phenotypically susceptible isolate was found to be resistant by genotypic identification (Table 4).
| MDRAB(n=5) | |||||||
| Age group | Samples | Gender | Total | Molecular identification using BLASTA tool | |||
| Male Female | |||||||
| 1- 20 | Urine | 1 | 1 | 2 | |||
| 21 - 40 | Pus | 1 | 1 | 2 | 5 isolates were dentified as Acinetobacter baumannii | ||
| 41- 60 | Sputum | 1 | 1 | ||||
| Total | 3 | 2 | 5 | ||||
| Susceptible AB(n=1) | |||||||
| Age group | Samples | Gender | Total | Molecular identification using BLASTA tool | |||
| Male Female | |||||||
| 21 – 40 | Pus | Nil | 1 | 1 | 1 isolate was identified as Acinetobacter baumannii and NCBI accession number (MW582860) | ||
Table 4: Association of age, gender and molecular identification using sequencing for 5 MDRAB and 1 susceptible AB among clinical samples (n=6).
Discussion
In the present study, the phenotypic assessment of virulence factor revealed 52 percent were positive for ESBL production, likewise, in a study 59% of the isolates were positive for ESBL. In other studies 27 percent and 27.5 percent of the isolates were positive for ESBL, which was lower than the present study had reported ESBL production to be 5.2 percent which is much lesser as compared to this study had reported ESBL production in A. baumannii as 83.6 percent, which is higher as compared to this study. In this study, 63 percent isolates of A. baumannii were positive for Carbapenamase and MBL production [27-29]. People have reported very high MBL production in A. baumannii as 96.6% and 80.3%, respectively, which was higher than the present study. Gupta had reported MBL production to be 7.5 percent which is much lesser as compared to this study. These variations in prevalence of various resistant enzymes could be due to difference in the antibiotic usage and judicious selection of antibiotics in their hospital settings. Hence this high rate of ESBL, MBL production by the clinical isolates of Acinetobacter baumannii warrants routine detection of beta lactamase enzymes which will enable the clinicians not only to prescribe appropriate treatment to patients. As per literature search, no other studies are available to indicate any correlation with ESBL, MBL production among MDRAB isolates. The molecular characterization was carried out on 15 MDRAB strains using conventional Polymerase Chain Reaction (PCR), DNA extraction and sequence analysis. Eight groups of resistant genes were identified. It was observed that blaOXA 51 and blaOXA23 genes were amplified for carbapenemase genes in 100 percent and 87 percent of the isolates respectively [30-33].
Further, blaTEM and blaCTX-M were amplified against ESBL gene in 20 percent and 13 percent of the isolates, while blaVIM and blaIMP were amplified against MBL gene in 53 percent and 40 percent of the isolates respectively. In a study done by Manohar et al. blaCTX-M was identified in 58.6 percent of the isolates, which was found to be higher than the present study revealed A. baumannii harboring blaOXA-51-like gene has been identified as a marker for species identification. An intrinsic blaOXA-51-like gene detected in all isolates in this study supports the use of this gene as a surrogate marker of A. baumannii identification. In another study done 93 percent of the isolates demonstrated the presence of blaOXA51-like genes, while blaIMP was present among 51 percent of the genes amplified against MBL, which is in concordance with the present study. In a study blaOXA-51-like gene and blaOXA 23-like were prevalent in 98 percent and 96 percent of the resistant strains respectively, similar to the present study. In contrast a study for the detection of ESBL genes showed that blaCTX-M gene was found amplified in 30% of the isolates, which was higher than the present study.
The first identification of OXA-48-producing Enterobacteriaceae was in an isolate in Turkey in 2001 and shortly thereafter, there was an outbreak of OXA-48- producing Enterobacteriaceae. Reported in Istanbul in 2006. In the present study, Carbapenamase OXA-48 gene was not detected in MDRAB. Among MBLgene, blaNDMlike genes was not detected in the current study, whereas other studies showed the presence of blaNDM-like gene. The genomic analysis among five isolates showed 100 percent positivity for Acinetobacter baumannii. The PCR products were sequenced and identified to the species level using the BLASTN tool. Unlike other studies, this study showed the presence of virulence gene blaoxa 51 in a susceptible isolate which reveals the importance of testing gene identification even in susceptible isolates.
Conclusion
Based on the genotypic evaluation, the study has demonstrated the expression of various genes encoding for Extended Spectrum β-Lactamase (ESBL), carbapenemase and Metalloβ-Lactamase (MBL). The most relevant finding of this study was the identification of blaOXA 51 gene in one of the 15 susceptible isolates isolated from pus samples from female patient. This indicates that susceptible isolates should also be tested for the presence of virulence genes. This will help in choosing the appropriate antibiotic for the treatment of AB infections.
Recommendation
This study indicates the need for testing of even susceptible isolates for virulence genes.
Acknowledgements
We would like to thank Dr.M Jayalakshmi (Mother), Dr. Sathyanaraya N Gummadi, IIT Madras and Dr. Ramesh N, VIT, Vellore and Dr. R Vijayaraghavan SIMATS for their guidance.
References
- Abdullahi IN, El-fulaty AA, Faruku N, et al. Clinical and epidemiological significance of RT-PCR and non-structural glycoprotein-1 assays in the diagnosis of dengue virus infections. Alexandria J Med 2020; 56:1-3.
- Kaur A, Singh S. Prevalence of Extended Spectrum Betalactamase (ESBL) and Metallo Beta Lactamase (MBL) producing Pseudomonas aeruginosa and Acinetobacter baumannii isolated from various clinical samples. J pathogens 2018.
- Bali EB, Acik L, Sultan N. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended-spectrum beta-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. Afr J Microbiol Res 2010; 4:650-654.
- Banerjee T, Mishra A, Das A, et al. High prevalence and endemicity of multidrug resistant Acinetobacter spp. in intensive care unit of a tertiary care hospital, Varanasi, India. J pathogens 2018.
- Bauer AW, Perry DM, Kirby WM. Single-disk antibiotic-sensitivity testing of staphylococci: An analysis of technique and results. Arch Intern Med 1959; 104:208-216.
- Bradford PA. Extended-spectrum ß-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 2001; 14:933-951.
- Chaudhary M, Payasi A. Molecular characterization and antimicrobial susceptibility study of Acinetobacter baumannii clinical isolates from Middle East, African and Indian patients. J Proteomics Bioinform 2012 ;5:265-269.
- Humphries RM, Ambler J, Mitchell SL, et al. On behalf of the clsi methods development and standardization working group of the subcommittee on antimicrobial susceptibility testing. 2018. CLSI Methods Development and Standardization Working Group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol 2018; 56:01934.
- Eze EC, Chenia HY, El Zowalaty ME. Acinetobacter baumannii biofilms: Effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect Drug Resist 2018; 11:2277.
- Ghaith DM, Zafer MM, Al-Agamy MH, et al. The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann Clin Microbiol 2017; 16:1-8.
- Gupta V, Datta P, Chander J. Prevalence of metallo-ß lactamase (MBL) producing Pseudomonas spp. and Acinetobacter spp. in a tertiary care hospital in India. J Infec 2006; 52:311-4.
- Gupta N, Gandham N, Jadhav S, et al. Isolation and identification of Acinetobacter species with special reference to antibiotic resistance. J Nat Sci Biol Med 2015; 6:159.
- Hodiwala A, Dhoke R, Urhekar AD. Incidence of metallo-betalactamase producing pseudomonas, acinetobacter & enterobacterial isolates in hospitalised patients. Int J Pharamcy Biol Sci 2013; 3:79-83.
- Hsu LY, Apisarnthanarak A, Khan E, et al. Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in south and southeast Asia. Clin Microbiol Rev 2017; 30:1-22.
- Jamal S, Al Atrouni A, Rafei R, et al. Molecular mechanisms of antimicrobial resistance in Acinetobacter baumannii, with a special focus on its epidemiology in Lebanon. J Glob Antimicrob Resist 2018; 15:154-63.
- Jiang W, Liu H, Zhong M, et al. Study on the resistant genes to carbapenems and epidemiological characterization of multidrug-resistant Acinetobacter baumannii isolates. Microb Drug Resist 2013; 19:117-23.
- Kaur A, Gupta V, Chhina D. Prevalence of metalo-ß-lactamase-producing (MBL) Acinetobacter species in a tertiary care hospital. Iran J Microbiol 2014; 6:22.
- Kuo, SC, Chang, SC, Wang, HY, et al. Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: nationwide data from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program. BMC 2012; 12:1-9.
- Li P, Niu W, Li H, et al. Rapid detection of Acinetobacter baumannii and molecular epidemiology of carbapenem-resistant A. baumannii in two comprehensive hospitals of Beijing, China. Front Microbiol 2015; 6:997.
- Lee CR, Lee JH, Park M, et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017; 7:55.
- Noyal MJC, Menezes GA, Harish BN, et al. Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative gram negative bacteria. Indian J Med Res 2009; 129:707-712.
- Oberoi L, Singh N, Sharma P et al. ESBL, MBL and Ampc beta Lactamases Producing Superbugs. Havoc in the Intensive Care Units of Punjab India. J Clin Diagn Res 2013; 7: 70-73.
- Palzkill T. Metallo-beta-lactamase structure and function. Ann N Y Acad Sc 2013; 1277: 91-104.
- Poirel L, Heritier C, Tolu¨n V, et al. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 2004; 48:15-22.
- Abd El-Baky RM, Farhan SM, Ibrahim RA, et al. Antimicrobial resistance pattern and molecular epidemiology of ESBL and MBL producing Acinetobacter baumannii isolated from hospitals in Minia, Egypt. Alexandria J Med 2020; 56:4-13.
- Saranathan R, Vasanth V, Vasanth T, et al. Emergence of carbapenem non-susceptible multidrug resistant Acinetobacter baumannii strains of clonal complexes 103B and 92B harboring OXA-type carbapenemases and metallo-ß-lactamases in Southern India. Microbiol Immunol 2015; 59:277-84.
- Vijayakumar S, Gopi R, Gunasekaran P, et al. Molecular characterization of invasive carbapenem-resistant Acinetobacter baumannii from a tertiary care hospital in South India. Infect Dis Ther 2016; 5:379-87.
- Stewart A, Harris P, Henderson A, et al. Treatment of infections by OXA-48-producing Enterobacteriaceae. Antimicrob Agents Chemother 2018; 62:e01195-1118.
- Traglia GM, Chua K, Centron D, et al. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol Evol 2014; 6:2235-2239.
- Vijayakumar S, Gopi R, Gunasekaran P, et al. Molecular characterization of invasive carbapenem-resistant Acinetobacter baumannii from a tertiary care hospital in South India. Infect Dis Ther 2016; 5:379-87.
- Wen Y, Ouyang Z, Yu Y, et al. Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region. Nucleic Acids Res 2017; 45: 9773 9787,
- Yang H, Huang L, Barnie PA, et al. Characterization and distribution of drug resistance associated ß lactamase, membrane porin and efflux pump genes in MDR A. baumannii isolated from Zhenjiang, China. Int J Clin Exp, Med 2015; 8: 15393 15402.
- Yong D, Lee K, Yum JH, et al. Imipenem EDTA disk method of differenciation of metallo beta lactamase producing clinical isolates Pseudomonas species and Acinetobacter species. J Clin Microbiol 2002; 40;3798-3801.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Author Info
Anitha M1*, Umamageswari S.S.M1 and Sumathi G2
1Department of Microbiology, Vels Medical College and Hospital, Vels University, Manjankaranai Village, Tiruvalur District, Chennai, India2Department of Microbiology, Sri Muthu Kumaran Medical College Hospital and Research Institute, Tamil Nadu Dr. M.G.R Medical University, Chikkarayapuram, Chennai, India
Citation: Anitha M, Prabha R, Sumathi G, Virulence Genes and Genomic Sequence of A. Baumannii from Different Clinical Samples, J Res Med Dent Sci, 2023, 11(4):17-22.
Received: 29-Mar-2023, Manuscript No. jrmds-23-94363; Accepted: 01-Apr-2023, Pre QC No. jrmds-23-94363; Editor assigned: 01-Apr-2023, Pre QC No. jrmds-23-94363; Reviewed: 15-Apr-2023, QC No. jrmds-23-94363; Revised: 22-Apr-2023, Manuscript No. jrmds-23-94363; Published: 29-Apr-2023
