Research - (2020) Volume 8, Issue 1
Upregulated CCL20 and CCR6 in Cancer Stem Cells Converted from Mouse iPS Cells
Juan Du1, Akimasa Seno1,2,3, Saki Sasada1, Yanning Xu1,4, Aung Ko Ko Oo1, Ghmkin Hassan1, Shunsuke Ueno2, Said M Afify1, Maram H Zahra1, Nobuhiro Okada2, Ling Chen4, Xiaoying Fu5, Heizo Tokutaka6, Ting Yan7 and Masaharu Seno1,2,3*
*Correspondence: Masaharu Seno, Department of Medical Bioengineering, Graduate School of Natural Science and Technology, Okayama University, Okayama, Japan, Email:
Abstract
Background: Cancer stem cells (CSCs) as a class of malignant cancer cells play an important role in tumor progression. Previous studies by our group have demonstrated the establishment of the model of CSCs converting mouse iPS cells (miPSCs) into CSCs by treating the miPSCs with a conditioned medium (CM) of Lewis Lung Carcinoma (LLC) cells with or without the nonmutagenic chemical compounds. CSCs converted from miPSCs developed highly malignant adenocarcinoma when subcutaneously transplanted into the nude mice.
Methods: The miPSCs were treated with each compound for 1 week in the presence of a CM of LLC cells. We evaluated the gene expression in the resultant CSCs comparing that in miPSCs by microarray analysis. And the expression of chemokine (C-C motif) ligand 20 (CCL20) and C-C chemokine receptor type 6 (CCR6) in converted cells were evaluated by rt-qPCR. The CCR6 expression in converted cells and primary cells were determined by flow cytometry.
Results: As the result, the expression of CCL20 was found upregulated in the presence of CM supplemented with PD0325901. Then we assessed the expression of CCR6, which was considered to be stimulated by CCL20. Then the expression of CCR6 was also found up-regulated. Interestingly, IL17A expression was also observed only in the CSCs from the primary tumor implying the effect of tumor microenvironment. Moreover, significantly high level of CCR6 was showed in flow cytometric analysis.
Conclusion: These results suggest that a model of CSCs with CCL20-CCR6 autocrine loop was obtained as the result of the conversion of iPSCs. This CSC should be a good model to study targeting CCR6 as a G protein-coupled receptor (GPCR).
Keywords
miPSCs, CSCs, CCR6, CCL20
Introduction
The CSCs has been identified as a subpopulation of stem-like cells within tumors exhibiting characteristics of both stem cells and cancer cells. CSCs are characterized by the abilities of self-renewal, differentiation in vitro and tumorigenesis in vivo [1]. CSCs are nowadays generally accepted to represent a unique population of cancer tissues that are tumorigenic, and resistant to most chemotherapeutic agents and radiation therapy, supporting cancer progression and recurrence.
The molecular and cellular mechanisms of cancer progression are now significantly being studied due to the finding of CSCs. Chronic inflammation is considered one of the reasons essential for cancer initiation and progression providing a tumor microenvironment. Chemokines are known to play a prominent role in inflammation and spreading of cancer. Therefore, chemokines and their cognate receptors should be the important factors involved in wound healing and angiogenesis. The ligands and their receptors are generally expressed in various cells in response to inflammation to recover homeostasis from imbalanced situation [2].
On the other hand, the chemokine CCL20 is considered a sole known ligand specific to CCR6, one of the GPCRs. Their interaction has been confirmed in B and T cells as well as dendritic cells (DCs) [3-5]. Recently, CCR6 has been shown in promoting non-small-cell-lung- cancer (NSCLC) carcinogenesis [6]. The chemokine/ chemokine receptor pair CCL20/CCR6 is a key player in lung immunity [7]. The interaction of CCL20 and CCR6 is thought to trigger the inflammation, which leads to cell chemotaxis [8,9]. Furthermore, recent research targeting the biological function of CCR6 indicates a significant role for this chemokine receptor in the development of NSCLC [10].
Previously, our group has demonstrated the development of CSCs from miPSCs treating with the CM derived various cancer cell lines [11-13]. The conditioned medium was supposed to be enriched with various factors related with the tumor microenvironment including inflammatory cytokines, chemokines and growth factors. In this study, we demonstrated the expression of CCL20 and CCR6 be upregulated in the presence of CM supplemented with PD0325901. And significantly high level of CCR6 was detected in flow cytometric analysis. The CCL20/CCR6 axis was involved in the progression of the conversion of CSCs. We suggested that a model of CSCs with CCL20-CCR6 autocrine loop was obtained as the result of the conversion of iPSCs. This CSC model should be a good model to study targeting CCR6 as a GPCR.
Materials and Methods
Materials
Mouse induced pluripotent stem cells (miPSCs, iPS-MEF-Ng-20D-17; Lot No.012) were provided from RIKEN Cell Bank, Japan. DMEM, 2-mercaptoethanol, collagenase, gelatin, Hematoxylin and Eosin Y were from Sigma, NY. KnockOutTM Serum Replacement (KSR) and Non-Essential Amino acids (NEAA) were from Gibco, NY. L-Glutamine, 2.5% Trypsin and CaCl2 were from Nacalai Tesque, Japan. 100 U/ ml penicillin/streptomycin (P/S) cocktail were from Wako, Japan. Leukemia inhibitory factor (LIF) was from Millipore, MA. Mitomycin C treated mouse embryonic fibroblasts (MEF) were provided from Reprocell, Japan. Mouse Lewis lung carcinoma (LLC) cells were obtained from ATCC, VA, USA. PD0325901 (391210-10-9) and CHIR99021 (252917-06-9) were purchased from Selleck Chemicals, Japan. Dasatinib (302962-49-8) was purchased from AdooQ BioScience, USA.
Anti-mouse CCR6 goat polyclonal antibody (NB100-713) was purchased from Novus Biologicals USA. Donkey polyclonal anti-Goat lgG-H&L (Alexa Fluor®647) antibody (ab150135) was purchased from Abcam, UK. Horseradish peroxidase (HRP)-conjugated anti-goat IgG donkey polyclonal antibody (sc-2020) was purchased from cell santa cruz biotechnology, Inc. Antimouse β-actin rabbit polyclonal antibody (4970), HRP-conjugated anti-rabbit IgG goat polyclonal antibody (7074) were purchased from Cell Signaling Technology, MA, USA.
Cell culture
miPSCs were maintained in DMEM supplemented with 15% FBS, 0.1mm NEAA, 2 mm L-Glutamine, 50 U/ml P/S, 0.1 mm 2-mercaptoethanol and 1000 U/ml of LIF on feeder layers of mitomycin C treated MEF. In the case of feeder-less, the miPSCs were cultured on gelatin coated dishes. LLC cells were maintained in DMEM containing 10% FBS supplemented with 100 U/ml P/S.
To prepare the CM from LLC cells, the medium was collected as previously described. The mixture of CM and miPS medium (1:1) was supplemented with each signal inhibitor. The miPSCs were cultured in the mixed medium without LIF and MEF feeder cells. One half of the medium was replaced with fresh mixed medium every day up to day 6. On day 7, the expression of GFP and cell morphology was observed and photographed using an inverted fluorescent microscope (Olympus IX81, Japan).
RNA extraction, cDNA synthesis and reverse transcription quantitative PCR analysis
Total RNA was extracted from cells using the RNA easy Mini kit (QUIAGEN, Germany) and treated with DNase according to the manufacturer's instructions and 1 μg of RNA was reverse transcribed using the Superscript First- Strand kit (Invitrogen, CA). Reverse transcription quantitative PCR (rt-qPCR) was performed with SYBR green I Master Mix (Roche, Switzerland) using LightCycler® 480 Instrument (Roche, Switzerland) according to manufacturer’s instructions (Table 1).
| Names | Accession | Forward Primer Sequence 5’- 3’ | Reverse Primer Sequence 5’- 3’ |
|---|---|---|---|
| CCL20 | NM_016960.2 | CGACTGTTGCCTCTCGTACA | GCTTCATCGGCCATCTGTCT |
| CCR6 | NM_001190334.1 | AGCTCAGCATTTTCTGGGCTTCA | GTGATGGGCTCTGAGACAGAC |
| IL17A | NM_010552 | GGGTCTTCATTGCGGTGGAGAG | ATCCCTCAAAGCTCAGCGTGTC |
| IL17A Receptor | NM_008359.2 | ACAGTTCCCAAGCCAGTTGC | TCAGCACGATGACAGATCCC |
Table 1: Primers used.
Microarray analysis of gene expression
Total RNA was extracted with QIAGEN RNeasy kit and isolated RNA was treated with DNase as described avove. Then the RNA was used as the template of cDNA, which was subjected to microarray analysis employing SurePrint G3 Mouse Gene Expression (Agilent, CA, USA).
Self-organizing map analysis
The spectrum data obtained from microarray was analyzed by SOM (SOM Japan Inc., Japan). The values of read from the hybridized spots were compared and the difference between the miPSCs treated with PD0325901 and the average values of miPSCs treated with or without ATRA in the presence of CM was calculated. SOM was used to find an ideally expressed gene, which was named as an ideal probe (IP), then the genes were clustered by SOM with the distance of each gene from the IP gene supposed to be expressed in miPSCs treated with PD0325901 but not in miPSCs treated with or without ATRA in the presence of CM.
Flow cytometry
The miPSCs that survived following the 1-week treatment were transferred to a 60-mm dish were harvested with a rubber scraper. The cells were then dissociated by passing through a mesh and incubated with anti-mouse CCR6 antibody (1:50) in 100μL for 30 min at 4°C Cells were washed with ice-cold PBS and incubated with the secondary antibody (1:500) in 100μL for 30 min at 4°C followed by wash with ice-cold PBS. The resultant cells were re-suspended in 100μL of ice-cold PBS and analyzed by a flow cytometer (BD AccuriTM C6 plus, Becton & Dickinson, NJ). Data from each experiment was analyzed by FlowJo software (FlowJo, LLC, Ashland, OR, USA).
Statistical analysis
The fluorescence data read by the microplate reader were analyzed using the two-tailed student’s t-test and presented as the mean ± standard deviation (SD) from independent experiments repeated at least three times. The statistical significance in mean values between two groups was determined by 2-tailed student’s t-test and expressed as the mean values. In the data acquired from the rt-qPCR analysis, the statistical significance between the mean values of more than two groups was determined using one-way analysis of variance (ANOVA) and Dunnett's multiple comparisons test. P<0.05 was considered statistically significant.
Results and Discussion
The effect of chemical compounds during the conversion of miPSCs into CSCs
Previously, we found the miPSCs converted into CSCs, exhibiting self-renewal, differentiation potential and tumorigenesis, in the presence of a CM of cancer derived cell lines in 4 weeks [11-13]. We employed miPSCs expressing a gene encoding green fluorescent protein (GFP) under the control of the Nanog promoter [14] to distinguish the undifferentiated/differentiated condition of the cells. In miPSCs, GFP fluorescence is kept on during the undifferentiated state, while GFP fluorescence is turned off when the miPSCs differentiated in the absence of leukemia inhibitory factor (LIF). Applying this procedure, in this study we evaluated the risk of non-mutagenic chemical compounds to accelerate the conversion, which was considered as CSSC induction. Most of the compounds assessed were the inhibitors of cytoplasmic signaling pathways. As the results, the expression of GFP fluorescence were obviously maintained after 1 week when miPSCs were cultured with PD0325901 in the presence of CM from LLC cells (Figure 1). In contrast, All-Trans Retinoic Acid (ATRA) significantly accelerated diminishing GFP fluorescence indicating the cells differentiated. In our recent study, we have demonstrated that three compounds, PD0325901, CHIR99021 and Dasatinib, out of 110 could accelerate the conversion of miPSCs into CSCs, and the expression of GFP fluorescence were maintained. PD0325901 is known as an inhibitor of MEK, and MEK is a main downstream of tyrosine kinases from Ras/Raf/MAP kinase cascade. Since Dasatinib is known as the tyrosine kinase inhibitor of src, bcl and c-kit, PD0325901 and Dasatinib appeared to have similar role in converting miPSCs into CSCs. When PD0325901 combined with CHIR99021 together the selfrenewal potential of mouse ESCs was maintained while CHIR99021 was an inhibitor of GSK-3β [15]. In this context, the functions of three inhibitors might closely be related one another to enhance the expression of GFP fluorescence and maintain the stemness resulting in the conversion from miPSCs into CSCs.
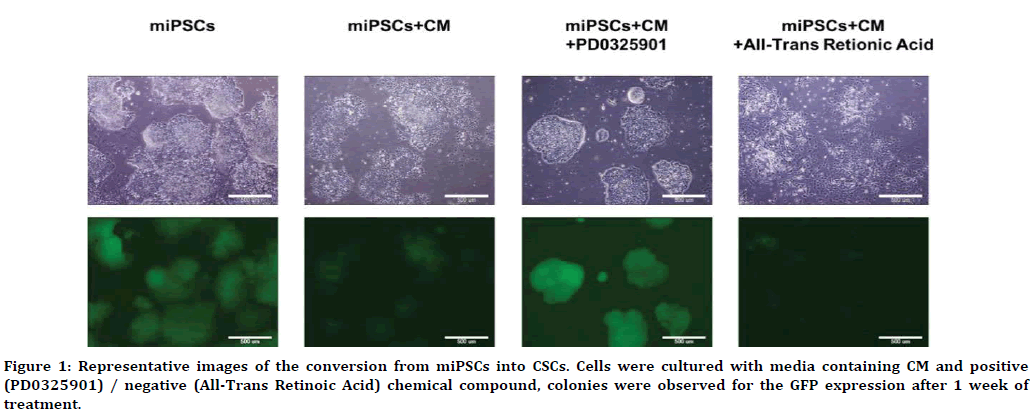
Figure 1. Representative images of the conversion from miPSCs into CSCs. Cells were cultured with media containing CM and positive (PD0325901) / negative (All-Trans Retinoic Acid) chemical compound, colonies were observed for the GFP expression after 1 week of treatment.
Gene expression difference during the conversion into CSCs treated with MEK inhibitor
We analyzed the difference of gene expression between the cells treated with PD0325901and ATRA by microarray followed by gene clustering analysis with sphere self-organizing map (sSOM) [16-18]. First of all, miPSCs and miPSCs treated with the CM was compared to reveal the genes which is related to CSC conversion. There were 507 up-regulated and 447 down-regulated genes (Figure2A). PD0325901 could have effects on them during the CSCs conversion. The expression of these 964 genes were further compared between the miPSCs treated with CM and PD0325901 and those treated with CM and ATRA. Then we found 69 genes upregulated and 320 gens downregulated by PD0325901 (Figure 2B). We supposed an IP, which was not expressed in miPSCs treated with CM or with CM and ATRA, but highly expressed in miPSCs treated with CM and PD0325901. The relationships of each gene and IP were analyzed by sSOM and depicted on the sphere (Figure 2C and 2D). The distances reflected on the sphere between each gene and the IP was sorted from the shortest to the longest and the a part of the shortest genes are shown in Table 2. The more effect should be on the gene by PD0325901 if the distance from the gene is closer to IP. From this point of view, we thought CCL20 could be the candidate to be analyzed for the relationship with CSCs and PD0325901 because CCL20 is known as a chemokine stimulating CCR6, which was a GPCR activating PI3K signaling pathway.
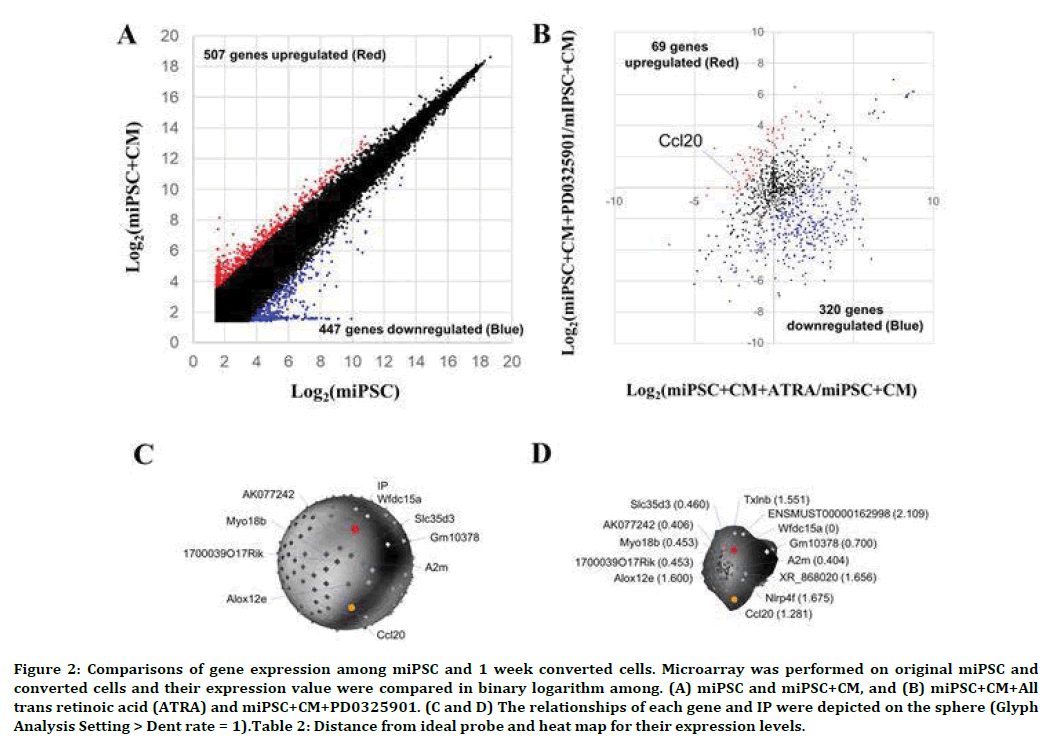
Figure 2. Comparisons of gene expression among miPSC and 1 week converted cells. Microarray was performed on original miPSC and converted cells and their expression value were compared in binary logarithm among. (A) miPSC and miPSC+CM, and (B) miPSC+CM+All trans retinoic acid (ATRA) and miPSC+CM+PD0325901. (C and D) The relationships of each gene and IP were depicted on the sphere (Glyph Analysis Setting > Dent rate = 1).Table 2: Distance from ideal probe and heat map for their expression levels.
| From IP | Agilent ID | a b c | Systematic Name | David ID | Name |
|---|---|---|---|---|---|
| 0 | A_51_P323770 |  |
NM_183271 | 68221 | WAP four-disulfide core domain 15A (Wfdc15a) |
| 0.404 | A_52_P455370 | NM_175628 | 232345 | alpha-2-macroglobulin (A2m) | |
| 0.406 | A_66_P101506 | AK077242 | |||
| 0.453 | A_55_P2728797 | AK018940 | 68208 | RIKEN cDNA 1700039O17 gene (1700039O17Rik) | |
| 0.453 | A_55_P2168383 | NM_028901 | 74376 | Myosin XVlllb (Myo18b) | |
| 0.46 | A_52_P467488 | NM_029529 | 76157 | Solute carrier family 35, member D3 (Slc35d3) | |
| 0.7 | A_55_P2731446 | XM_011245278 | 101055806 | Predicted gene 10378 (Gm10378) | |
| 1.281 | A_51_P408595 | NM_016960 | 20297 | Chemokine (C-C motif) ligand 20 (CCL20) | |
| 1.551 | A_51_P315391 | NM_138628 | 378431 | Taxilin beta (Txlnb) | |
| 1.6 | A_51_P471659 | NM_145684 | 11685 | Arachidonate lipoxygenase, epidermal (Alox12e) | |
| 1.656 | A_55_P2900459 | XR_868020 | |||
| 1.675 | A_55_P1955871 | NM_175290 | 97895 | NLR family, pyrin domain containing 4F (Nlrp4f) | |
| 1.675 | A_51_P208145 | NM_021882 | 20431 | Premelanosome protein (Pmel) | |
| 1.767 | A_55_P2153021 | NM_011652 | 22138 | Titin (Ttn) | |
| 1.767 | A_52_P306007 | NM_024271 | 76413 | RIKEN cDNA 1700016D06 gene (1700016D06Rik) | |
| 1.858 | A_51_P291227 | NM_001005508 | 226652 | Rho GTPase activating protein 30 (Arhgap30) | |
| 1.858 | A_55_P2031999 | NM_145448 | 217830 | RIKEN cDNA 9030617O03 gene (9030617O03Rik) | |
| *The maximum distance from IP is 4.222, and genes whose distance is less than 2 is shown in this list. (A: miPSC+CM, B: miPSC+CM+PD0325901, C: miPSC+CM+ARTA) |
|||||
Table 2: Distance from ideal probe and heat map for their expression levels.
Expression of CCL20 and CCR6 in converted cells and primary cultured cells was upregulated
We further analyzed the expression of CCR6 and CCL20 in miPSCs treated with PD0325901, CHIR99021 and Dasatinib, using rt-qPCR. The expression level of CCR6 and CCL20 were found to be up-regulated during the 7 days of treatment (Figure 3A). Our previous work demonstrated that the miPSCs treated with the three compounds exhibited tumorigenicity when subcutaneously transplanted into Balb/c nude mice. Meanwhile, we evaluated the expression of CCR6 and CCL20 in the primary cells derived from the tumors formed in nude mice (Figure 3B). As the result, the significantly highly expression of CCR6 as well as CCL20 was found in the primary cells.
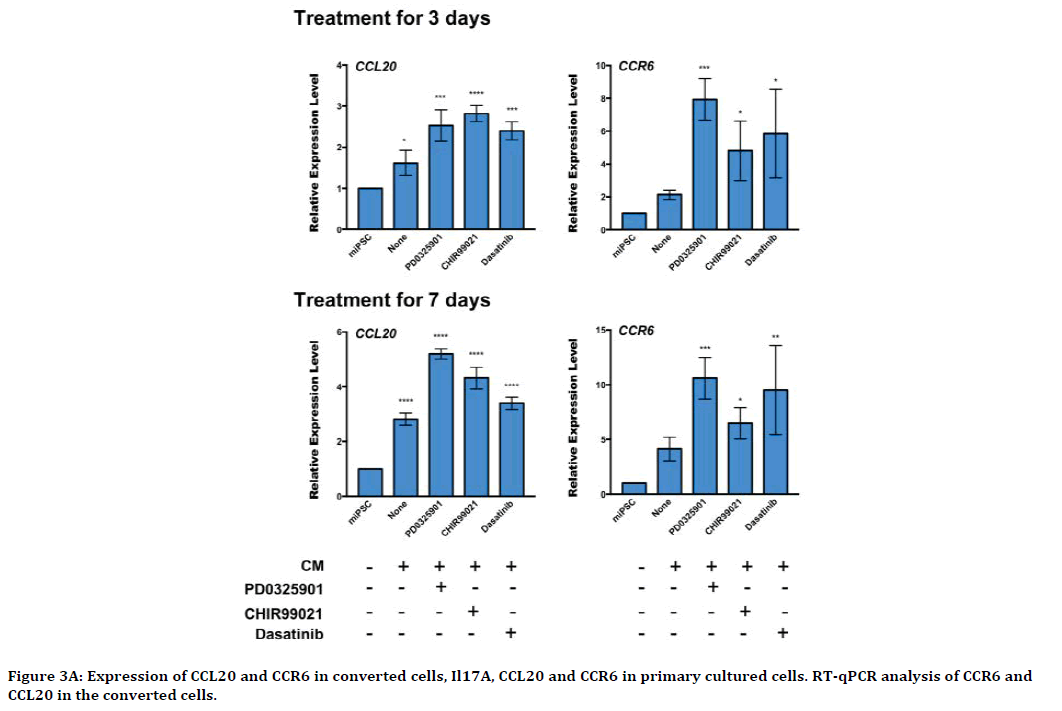
Figure 3A. Expression of CCL20 and CCR6 in converted cells, Il17A, CCL20 and CCR6 in primary cultured cells. RT-qPCR analysis of CCR6 and CCL20 in the converted cells.
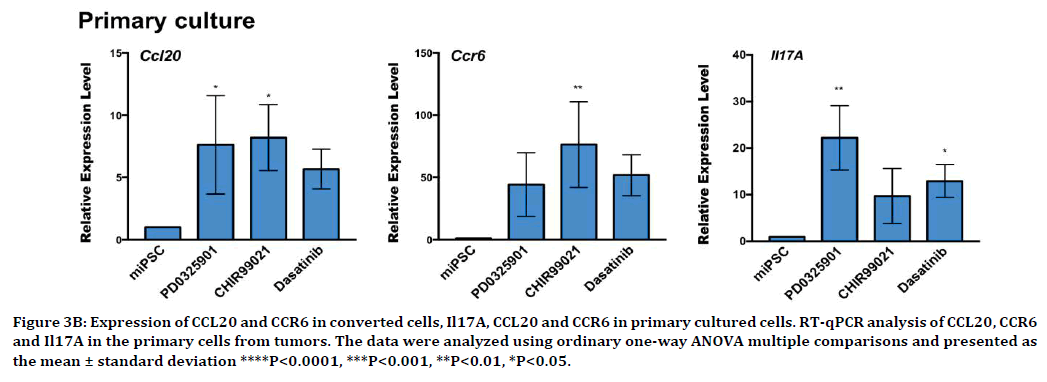
Figure 3B. Expression of CCL20 and CCR6 in converted cells, Il17A, CCL20 and CCR6 in primary cultured cells. RT-qPCR analysis of CCL20, CCR6 and Il17A in the primary cells from tumors. The data were analyzed using ordinary one-way ANOVA multiple comparisons and presented as the mean ± standard deviation ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05.
Expression of IL17A in primary cultured cells was detected
Recent studies have reported that IL17A, of which expression was driven by TGF-β and IL-6, induced CCL20 expression via the transcription factor STAT3 resulting in promoting tumor progression, and metastasis by upregulating the expression of the genes were involved in antiapoptotic, growth factors and angiogenesis [19-21]. However, these events were considered to be iunduced T lymphocytes. It may be explained by the presence of tumor associated macrophage (TAM) expressing the CCL20 surrounding the CSCs. We could not find the expression of IL17A in CSCs just after conversion of miPSCs while the slight upregulation of IL17A expression was detected in the primary cells from the tumor (Figure 3B). In the tumor microenvironment (TME), continuous exposure to CCL20 may have induced the expression of CCL20 in CSCs establishing autocrine stimulation of CCR on the CSCs transducing the signal to PI3K pathway. The expression of IL17A in the primary cells possibly implies the stimulation of the TAM to enrich the CCL20 in the TME although the significant expression of IL17AR was not observed in both CSCs just after conversion and those from primary tumor.
The inhibitors enhanced the expression of CCR6
Upregulation of the CCR6 expression was essentially related with organ orientation and significantly related with the metastasis of lung cancer [22-24]. We further evaluated the expression of CCR6 by flow cytometry and western blot. In flow cytometry, the CCR6-postive population in the presence of CM was found 41.1%, 21.7% and 31.6% in the cells when treated with PD0325901, CHIR99021 and Dasatinib, respectively while 27.4% in miPSCs. CCR6-postive population was 3.9% of miPSCs in the absence of the CM (Figure 4A). In the primary cells from the tumors, CCR6- postive population was significantly increased to 53.4%, 83.1% and 61.5% in the cells derived from primary tumors, primary-miPSCs+CM+PD0325901, primary-miPSCs+CM+CHIR99021 and primarymiPSCs+ CM+Dasatinib, respectively (Figure 4B). CCR6-postive population was 10.6% of primarymiPSCs cells.
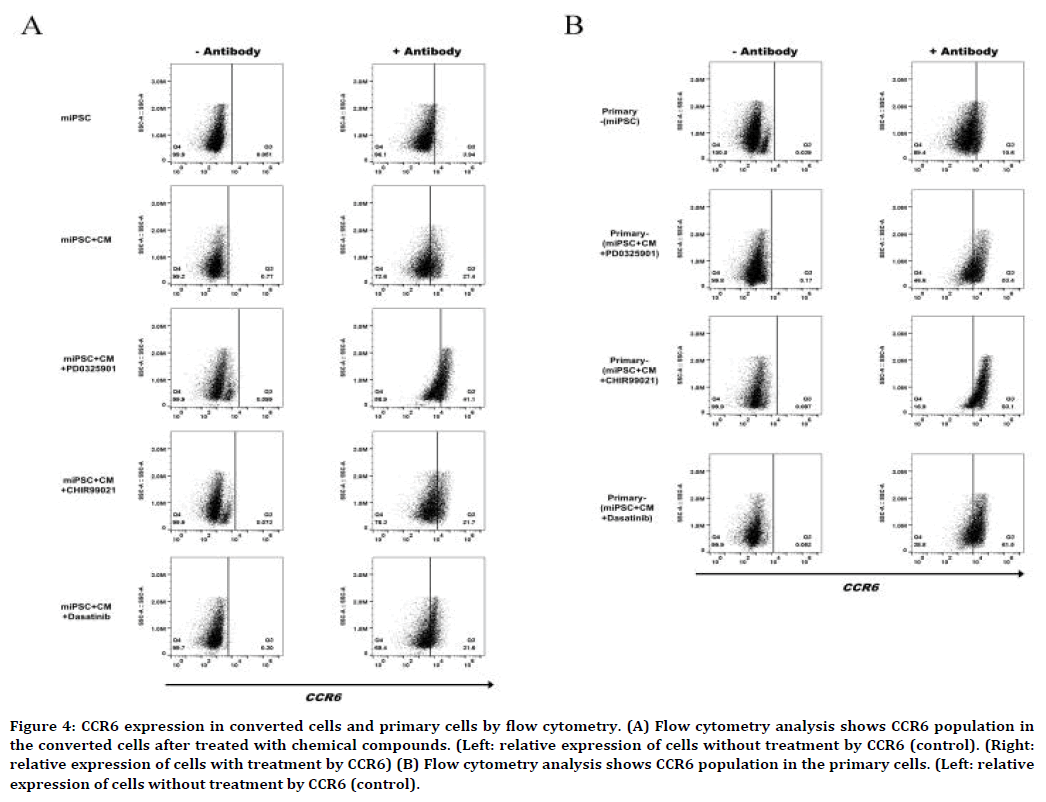
Figure 4. CCR6 expression in converted cells and primary cells by flow cytometry. (A) Flow cytometry analysis shows CCR6 population in the converted cells after treated with chemical compounds. (Left: relative expression of cells without treatment by CCR6 (control). (Right: relative expression of cells with treatment by CCR6) (B) Flow cytometry analysis shows CCR6 population in the primary cells. (Left: relative expression of cells without treatment by CCR6 (control).
Conclusion
In conclusion, our study demonstrated the expression of CCL20 and CCR6 be upregulated in the presence of CM supplemented with PD0325901. And significantly high level of CCR6 was detected in flow cytometric analysis. The CCL20/CCR6 axis was involved in the progression of the conversion of CSCs. We suggested that a model of CSCs with CCL20- CCR6 autocrine loop was obtained as the result of the conversion of iPSCs. This CSC model should be a good model to study targeting CCR6 as a GPCR.
Acknowledgments
This research study was partly supported by the Long-range Research Initiative No. 13 S01- 01-4 (MS), Japan Chemical Industry Association, and by JSPS Grant-in-Aid for Scientific Research (A) No. JP25242045 (MS), for Challenging- Exploratory Research No. JP26640079 (MS) and for Young Scientists (B) No. JP18K-15243 (AS). We are grateful for the scholarship of China Scholarship Council No. 201708050145 awarded for JD.
Authors Contribution
Masaharu Seno directed the whole project of study. Juan Du and Masaharu Seno designed the experiments. Juan Du, Akimasa Seno, Saki Sasada, Shunsuke Ueno and Yanning Xu conducted the experiments. Juan Du, Akimasa Seno, Masaharu Seno and Heizo Tokutaka analyzed the data. Aung Ko Ko Oo, Ghmkin Hassan, Said M. Afify and Maram H. Zahra supplied materials and supported analysis. Juan Du, Masaharu Seno, Nobuhiro Okada, Ling Chen, Ting Yan and Xiaoying Fu participated in the discussion. Juan Du and Masaharu Seno wrote the manuscript. All authors were involved in the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science 2009; 324:1670-1673.
- Sallusto F, Baggiolini M. Chemokines and leukocyte traffic. Nat Immunol 2008; 9:949-952.
- Varona R, Zaballos A, Gutierrez J, et al. Molecular cloning, functional characterization and mRNA expression analysis of the murine chemokine receptor CCR6 and its specific ligand MIP-3alpha. FEBS Lett 1998; 440:188-194.
- Liao F, Alderson R, Su J, et al. STRL22 is a receptor for the CC chemokine MIP-3alpha. Biochem Biophys Res Commun 1997; 236:212-217.
- Baba M, Imai T, Nishimura M, et al. Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J Biol Chem 1997; 272:14893-14898.
- Starner TD, Barker CK, Jia HP, et al. CCL20 is an inducible product of human airway epithelia with innate immune properties. Am J Respir Cell Mol Biol 2003; 29:627-633.
- Kirshberg S, Izhar U, Amir G. Involvement of CCR6/CCL20/IL-17 Axis in NSCLC disease progression. PLoS One 2011; 6:e24856.
- Annunziato F, Cosmi L, Santarlasci V, et al. Phenotypic and functional features of human Th17 cells. J Exp Med 2007; 204:1849-1861.
- Chen Z, O’Shea JJ. Th17 cells: A new fate for differentiating helper T cells. Immunol Res 2008; 41:87-102.
- Kirshberg S, Izhar U, Amir G, et al. Involvement of CCR6/CCL20/IL-17 Axis in NSCLC Disease Progression. PLoS one. 2011; 9:e24856.
- Chen L, Kasai T, Li Y, et al. A model of cancer stem cells derived from mouse induced pluripotent stem cells. PLoS One 2012; 7:e33544.
- Calle AS, Nair N, Seno M, et al. A new PDAC mouse model originated from iPSCs-converted pancreatic cancer stem cells (CSCcm). Am J Cancer Res 2016; 6:2799-2815.
- Nair N, Calle AS, Zahra MH, et al. A cancer stem cell model as the point of origin of cancer-associated fibroblasts in tumor microenvironment. Sci Rep 2017; 7:6838.
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 2007; 448:313-317.
- Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature 2008; 453:519-523.
- Tuoya, Sugii Y, Satoh H, et al. Spherical self-organizing map as a helpful tool to identify category-specific cell surface markers. Biochem Biophys Res Commun 2008; 376:414-418.
- Seno A, Maruhashi T, Kaifu T, et al. Exacerbation of experimental autoimmune encephalomyelitis in mice deficient for DCIR, an inhibitory C-type lectin receptor. Exp Anim 2015; 64:109-119.
- Seno A, Kasai T, Ikenda M, et al. Characterization of gene expression patterns among artificially developed cancer stem cells using spherical self-organizing map. Cancer Inform 2016; 15:163-178.
- Homey B, Dieu-Nosjean MC, Wiesenborn A, et al. Up-regulation of macrophage inflammatory protein-3 alpha/CCL20 and CC chemokine receptor 6 in psoriasis. J Immunol 2000; 164:6621-6632.
- Pan B, Che D, Cao J, et al. Interleukin-17 levels correlate with poor prognosis and vascular endothelial growth factor concentration in the serum of patients with non-small cell lung cancer. Biomarkers 2015; 20:232-239.
- Pan B, Shen J, Cao J, et al. Interleukin-17 promotes angiogenesis by stimulating VEGF production of cancer cells via the STAT3/GIV signaling pathway in non-small-cell lung cancer. Sci Rep 2015; 5:16053-16066.
- Raynaud CM, Mercier O, Dartevelle P, et al. Expression of chemokine receptor CCR6 as a molecular determinant of adrenal metastatic relapse in patients with primary lung cancer. Clinic Lung Cancer 2010; 11:187-191.
- Sutherland A, Mirjolet JF, Maho A, et al. Expression of the chemokine receptor CCR6 in the Lewis lung carcinoma (LLC) cell line reduces its metastatic potential in vivo. Cancer Gene Ther 2007; 14:847-857.
- Ghadjar P, Rubie C, Aebersold DM, et al. The chemokine CCL20 and its receptor CCR6 in human malignancy with focus on colorectal cancer. Int J Cancer 2009; 125:741-745.
Author Info
Juan Du1, Akimasa Seno1,2,3, Saki Sasada1, Yanning Xu1,4, Aung Ko Ko Oo1, Ghmkin Hassan1, Shunsuke Ueno2, Said M Afify1, Maram H Zahra1, Nobuhiro Okada2, Ling Chen4, Xiaoying Fu5, Heizo Tokutaka6, Ting Yan7 and Masaharu Seno1,2,3*
1Department of Medical Bioengineering, Graduate School of Natural Science and Technology, Okayama University, Okayama, Japan2Laboratory of Nano-Biotechnology, Graduate School of Interdisciplinary Science and Engineering in Health Systems, Okayama University, Okayama 700-8530, Japan
3Okayama University Research Laboratory of Stem Cell Engineering in Detroit, IBio, Wayne State University, MI 48202, USA
4Department of Pathology, Tianjin Central Hospital of Gynecology Obstetrics, Tianjin, P.R. China
5School of Integrative Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin, P.R. China
6SOM Japan Inc, Tottori, Japan
7The Hong Kong University of Science and Technology Medical Center, Shenzhen Peking University, Shenzhen, P.R. China
Citation: Juan Du, Akimasa Seno, Saki Sasada, Yanning Xu, Aung Ko Ko Oo, Ghmkin Hassan, Shunsuke Ueno, Said M. Afify, Maram H Zahra, Nobuhiro Okada, Ling Chen, Xiaoying Fu, Heizo Tokutaka, Ting Yan, Masaharu Seno, Upregulated CCL20 and CCR6 in cancer stem cells converted from mouse iPS cells, J Res Med Dent Sci, 2020, 8(1):200-207.
Received: 02-Jan-2020 Accepted: 02-Dec-2020
