Research - (2020) Advances in Dental Surgery
Investigation of TNF-α, LT-α and TNFR-2 gene Polymorphisms in Patients with Periodontitis in Turkey
Selin Özkan-Kotiloğlu1, Gülcan Kuyucuklu2, Esra Baltacioglu3*, Güliz Güncü4, Erhan Dursun4, Erkan Sukuroglu3, Erdem Karabulut5, Emine Sibel Sümer6 and Ferda Alev Akal?n4
*Correspondence: Esra Baltacioglu, Department of Periodontology, Faculty of Dentistry, Karadeniz Technical University, Trabzon, Turkey,
Abstract
Introduction: One of the cytokines involved in periodontal breakdown in periodontitis is; Tumor necrosis factor (TNF), with two types; tumor necrosis factor-α (TNF-α) and lymphotoxin-α (LT-α). Aim: In the present study, the individual frequency of TNF-α -308 promoter, LT-α +252 intron, and TNF receptor-2 (TNFR-2) +587 exon polymorphisms associated with chronic (CP) and generalized aggressive periodontitis (GAP) were evaluated in Turkish population. Materials and Methods: This research was conducted on 224 periodontitis patients (P) (119 CP, 105 GAP patients) and 85 healthy controls (C). Blood samples were collected, and specific DNA regions were amplified by PCR method. Investigated gene polymorphisms were determined by RFLP (Restriction Fragment Length Polymorphism). Statistical analyses, including linkage analyses were performed. Results: While there was no statistically significant difference in LT-α +252 polymorphisms between the patients and controls, significant associations between TNF-α -308 GA genotype and CP and GAP were determined. For TNFR-2 (+587) T/G polymorphism; TT, TG and GG genotype and allele frequencies were analysed and significant differences were found in genotype and allele frequencies between the control and CP and GAP groups (p<0,001). Conclusion: This is the first study investigating the TNF-α, LT-α and TNFR-2 polymorphisms in CP and GAP in Turkish population. Significant differences were found in genotype and allele frequencies, in CP and GAP groups.
Keywords
Gene polymorphisms, Periodontitis, Tumor necrosis factor alpha, Lymphotoxin alpha, Tumor necrosis factor receptor 2
Introduction
Periodontitis is a destructive inflammatory disease of the supporting structures of teeth. It causes irreversible destruction of alveolar bone, loss of attachment fibres and tooth loss. It is a multifactorial disease due to its effecting factors such as genetic, environmental and microbiological. Although the occurrence of bacterial infection in the gingival sulcus initiates the periodontal disease, bacterial accumulation is not a direct cause of this tissue destruction, it arises from the interaction between bacteria and host immune response mechanism. The disease is triggered by the periodontal pathogens by inducing the inflammatory cascade which leads to tissue destruction [1].
It is reported that cytokines are responsible for various biological activities such as homeostasis, development, inflammation and repair. Since they are important messenger molecules, control of their secretion and functions are complicated and irregular expression of cytokines induces pathology [1]. It is known that polymorphisms in cytokine genes may change the expression levels of the cytokines [2].
There are several studies on the association between the expressions and the genetic polymorphisms of cytokines such as interleukin-1 (IL-1), tumor necrosis factor alpha (TNF-α), and proteases and structural molecules such as human leukocyte antigens (HLAs), matrix metalloproteinases (MMPs), catepsin-C and vitamin D and the induction and the development of periodontal disease [3,4].
Tumor necrosis factor alpha (TNF-α) and lymphotoxin alpha (LT-α) are proinflammatory cytokines and play critical roles in bone resorption and collegenase secretion which are characteristics of periodontal tissue destruction. TNF protein has two cell surface receptors namely TNF receptor-1 (TNFR-1) and TNF receptor-2 (TNFR-2). Both receptors are expressed in almost all cell types including macrophages, lymphocytes, neutrophils, and fibroblasts [5].
TNF-α and LT-α cause the lysis of tumor cells and they are secreted from macrophages and lymphocytes, respectively [6,7]. The TNF-α and LT-α genes are located on the long arm of the human chromosome 6 [8]. The region where they are placed is HLA class III, which contains the genes related to immune response [9]. This region is highly polymorphic and about 16 polymorphic areas were determined on and around TNF-α and LT-α genes [10].
There are several researches which have demonstrated that periodontitis has a genetic background [3]. While the possibility of the disease increases in the presence of genetic risk factors, absence of genetic factors reduces the probability [3]. Recent studies have focused on polymorphisms of genes encoded cytokines that may be used as genetic markers [11].
It is crucial to reveal the relationship between periodontitis and TNF-α, LT-α, TNFR-2 gene polymorphisms not only because of their functions, but also the highly polymorphic genomic regions. Therefore, the aim of this study is to investigate the association between periodontitis and TNF-α -308 promoter (rs1800629), LT-α +252 intron (rs909253) and TNFR-2 +587 exon (rs1061622) polymorphisms in Turkish population.
Materials and Methods
Clinical studies
Study groups
A total of 224 patients, including 119 chronic periodontitis (CP) patients, 105 generalized aggressive periodontitis (GAP) patients and 85 periodontally healthy control (C) individuals participated in this study. The participants were selected among individuals who were referred to Hacettepe University (Ankara-Turkey) and Karadeniz Technical University (Trabzon- Turkey), Faculty of Dentistry, Department of Periodoontology for their periodontal treatments or routine periodontal controls. The demographic data of the participants are demonstrated in Table 1. All of the individuals included in the study had the following criteria: 1) had no history of systemic disease; 2) had received no periodontal treatment; 3) had not taken antibiotics, anti-inflammatory drugs, or any other drugs during the past 3 months. The study protocol was approved by the Ethics Committees of Hacettepe University and Karadeniz Technical University. (Approval numbers; Hacettepe University: TBK 05/15-17, Karadeniz Technical University: B.30.2. KTU. 0.01.00.01/534). All patients were informed about the research and they completed a questionnaire to provide information about the history of their systemic conditions and smoking habits. Each participant was evaluated according to the personal and family medical and dental history and dental examinations.
| CP | GAP | Controls | |
|---|---|---|---|
| Gender, n (%)1 | |||
| Female | 56 (47) | 71 (68) | 57 (67) |
| Male | 63 (53) | 34 (32) | 28 (33) |
| Age (mean ± SD)2 | 39.5 ± 10.3 | 31.2 ± 7.7 | 25.3 ± 4.8 |
| Smoking status n (%)3 | |||
| Smoker | 35 (29) | 23 (22) | 10 (12) |
| Non-smokers | 84 (71) | 81 (78) | 75 (88) |
| PD (mm)4 | 3.9 ± 0.9 | 4.8 ± 1.3 | 1.4 ± 0.4 |
| CAL (mm)5 | 4.7 ± 1.2 | 5.5 ± 1.6 | 1.5 ± 0.4 |
| GI (mm)6 | 1.8 ± 0.5 | 2.0 ± 0.6 | 0.2 ± 0.4 |
| BOP (%)7 | 83.9 ± 22.6 | 93.2 ± 14.9 | 5.1 ± 1.2 |
| PI (mm)8 | 1.6 ± 0.7 | 1.6 ± 0.8 | 0.1 ± 0.1 |
| 1Gender: p=0.005 for CP vs Controls; p>0.05 for GAP vs Controls; p=0.002 for CP vs. GAP | |||
| 2Age: p=0.000 for CP vs Controls; p=0.000 for GAP vs Controls; p=0.002 for CP vs. GAP | |||
| 3Smoking status: p=0.002 for CP vs Controls; p>0.05 for GAP vs Controls; p>0.05 for CP vs. GAP | |||
| 4PD: p=0.000 for CP vs Controls; p=0.000 for GAP vs Controls; p=0.000 for CP vs. GAP | |||
| 5CAL: p=0.000 for CP vs Controls; p=0.000 for GAP vs Controls; p=0.000 for CP vs GAP | |||
| 6GI: p=0.000 for CP vs Controls; p=0.000 for GAP vs Controls; p=0.000 for CP vs GAP | |||
| 7BOP (%): p=0.000 for CP vs Controls; p=0.000 for GAP vs Controls; p=0.000 for CP vs GAP | |||
| 8PI: p=0.000 for CP vs Controls; p=0.000 for GAP vs Controls; p>0.05 for CP vs GAP | |||
Table 1: Demographic and clinical periodontal data of individuals in chronic periodontitis (CP), generalized aggressive periodontitis (GAP) and control groups.
Clinical periodontal examinations
The patients were clinically and radiographically diagnosed for CP or GAP according to the criteria accepted by the American Academy of Periodontology in 1999 [12]. The periodontal status of all individuals was determined by measuring periodontal probing depth (PPD) and clinical attachment level (CAL) (Williams Periodontal Probe, Hu Friedy, Chicago Illinois), Gingival Index (GI) [13], bleeding on probing (BOP) and Plaque Index (PI) [14] values. Periodontal bone levels were determined by evaluating the panoramic radiographes of the individuals.
The control group consisted of periodontally healthy individuals with no history of periodontal disease, with PPD ≤ 3mm; and CAL ≤ 1mm; GI score 0.
Laboratory studies
Isolation of genomic DNA
All blood samples of the individuals were collected in sterile 10ml tubes containing EDTA solution and they were stored at +4°C until processing. Genomic DNA isolation from whole blood was performed by using QIAamp Blood Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. The integrity of DNA samples were checked on 1% agarose gels and isolated DNAs were stored at -20°C until analysed for TNF-α -308, LT-α +252 and TNFR-2 +587 genotypes.
Genotyping
Polymorphisms were assessed by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) analysis. PCR amplifications were performed using sequence specific primers for the amplification of target regions (TPersonal Thermal Cycler, Biometra, Germany). PCR reaction was conducted in 50μl of total volume, containing 2μl of DNA template (approximately 200ng), 10 μM of each primer, dNTPs, 5μl of 10XBuffer and 1 unit of Hotstart Taq DNA polymerase (Qiagen, Germany). The 117 base pair regions of TNF-α gene, the 368 base pair regions of LT-α gene and 242 bp region of TNFR-2 gene were amplified. The size of amplicons was analysed by using 1.5% agarose gel with ethidium bromide staining. RFLP method was used for detection of polymorphisms. PCR products of both genes were digested by restriction enzyme NcoI (Promega). Primer sequences, PCR and restriction conditions and products were demonstrated in Table 2.
| Polymorphisms | Primer Sequences and PCR Conditions | Restriction Enzyme and Conditions | Products |
|---|---|---|---|
| LT-α +252 (A/G) | (F): 5' CTC CTG CAC CTG CTG CCT GGA TC 3' | NcoI | AA genotype: 368bp |
| (R): 5' GAA GAG ACG TTC AGG TGG TGT CAT 3' | 37°C, 16h | AG genotype: 368+235+133bp | |
| 94°C 3 min; (94°C 30s, 65°C 30s, 72°C 30s) X 32 cycles; 72°C 5 min. | GG genotype: 235+133bp | ||
| TNF-α -308 (G/A) | (F): 5' AGG CAA TAG GTT TTG AGG GCC AT 3' | NcoI | GG genotype: 97+20bp |
| (R): 5' ACA CTC CCC ATC CTC CCG GCT 3' | 37°C, 16h | GA genotype: 117+97+20bp | |
| 95°C 15 min; (94°C 1min, 60°C 1 min, 72°C 1min) X 35 cycles; 72°C 10 min. | AA genotype: 117bp | ||
| TNFR-2 +587 (T/G) | (F): 5' ACT CTC CTA TCC TGC CTG CT 3' | Hin1II (NlaIII) | TT genotype: 242bp |
| (R): 5' TTC TGG AGT TGG CTG CGT GT 3' | 37°C, 16h | TG genotype: 242+133+109bp | |
| 95°C 10 min; (94°C 1min, 56°C 1 min, 72°C 1min) X 35 cycles; 72°C 7 min. | GG genotype: 133+109bp |
Table 2: Primer sequences, restriction enzyme and expected product sizes for each polymorphism.
Restriction digestion products were determined electrophoretically on 3% agarose gel with Sybr Safe DNA stain (Invitrogen). The sequence information of primers, conditions of PCR and RFLP analysis and the sizes of expected products are shown in Table 2. Results obtaining from RFLP analysis were shown in Figures 1-3 for TNF-α, LT-α and TNFR-2 genotypes, respectively.
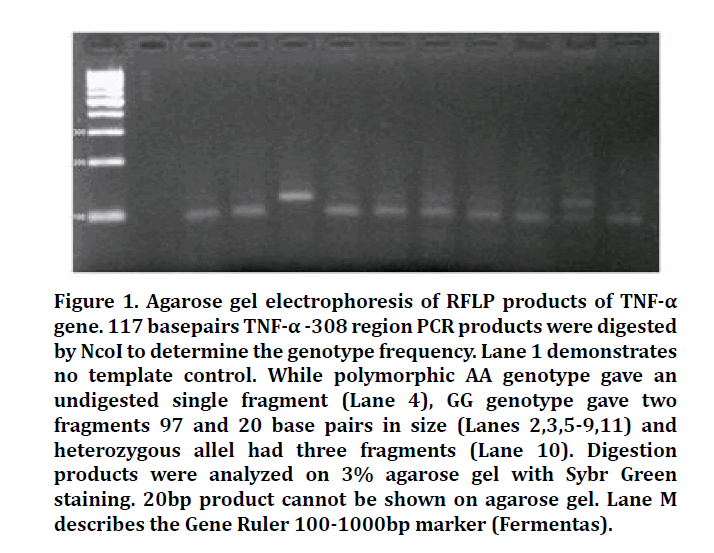
Figure 1: Agarose gel electrophoresis of RFLP products of TNF-α gene. 117 basepairs TNF-α -308 region PCR products were digested by NcoI to determine the genotype frequency. Lane 1 demonstrates no template control. While polymorphic AA genotype gave an undigested single fragment (Lane 4), GG genotype gave two fragments 97 and 20 base pairs in size (Lanes 2,3,5-9,11) and heterozygous allel had three fragments (Lane 10). Digestion products were analyzed on 3% agarose gel with Sybr Green staining. 20bp product cannot be shown on agarose gel. Lane M describes the Gene Ruler 100-1000bp marker (Fermentas).
Figure 2: Genotype profiles of LT-α +252 polymorphism on agarose gel. Polymorphic LT-α homozygous GG genotype has two fragments after NcoI digestion (Lanes 3-7) whereas homozygous AA genotype has one undigested band 368bp in size (Lanes 6 and 10). Heterozygous genotype has 3 different fragments (Lanes 4,5 and 8). . Lane M describes the Gene Ruler 100-1000bp marker (Fermentas).
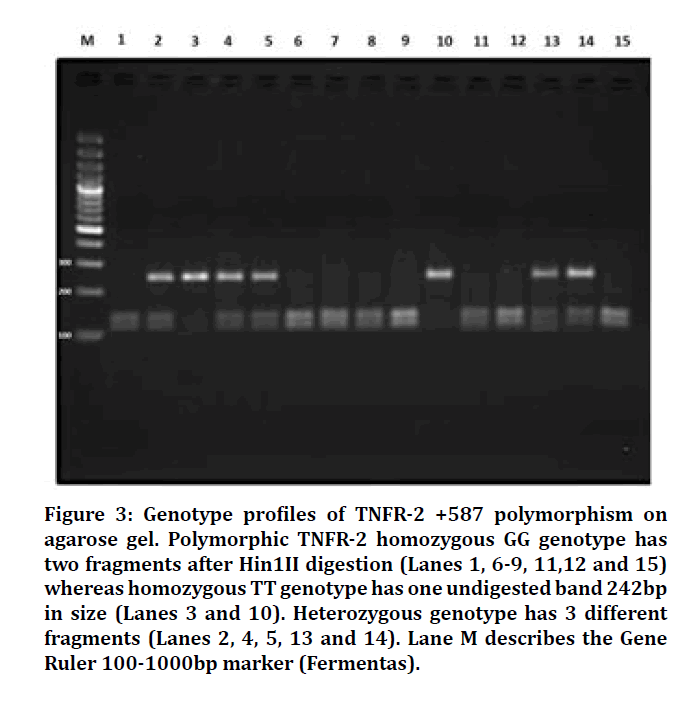
Figure 3: Genotype profiles of TNFR-2 +587 polymorphism on agarose gel. Polymorphic TNFR-2 homozygous GG genotype has two fragments after Hin1II digestion (Lanes 1, 6-9, 11,12 and 15) whereas homozygous TT genotype has one undigested band 242bp in size (Lanes 3 and 10). Heterozygous genotype has 3 different fragments (Lanes 2, 4, 5, 13 and 14). Lane M describes the Gene Ruler 100-1000bp marker (Fermentas).
Statistical analyses
Data analyses were carried out by using the IBM SPSS Statistics 23.0 version (IBM Corp., Armonk, NY). The χ² test and Mann-Whitney U test were used to examine the differences in distribution of demographic findings and clinical parameters, respectively. The χ² test was used to examine the differences in distribution of TNF-α, LT-α and TNFR-2 genotypes and allele frequencies in patients with two types of periodontitis and the referent population. p-Values of ≤ 0.05 were accepted as statistically significant. For Linkage Disequilibrium (LD), SNPassoc package in R software was used.
Results
Demographic and clinical findings
All individuals involved in the study were evaluated according to their age, gender, and smoking conditions (Table 1). Clinical periodontal parameters of the individuals and the comparisons of the groups are demonstrated in Table 1. PD, CAL, GI, %BOP and PI scores of GAP patients were significantly higher when compared to control group (p<0.000). In addition, all clinical parameters of CP patients were significantly higher when compared to control group (p<0.000). While PD, CAL, GI and %BOP scores of GAP patients were significantly higher when compared to CP patients (p<0.000), PI score showed no significant difference between those groups (p>0.05).
Laboratory findings
TNF-α-308: Results showed that periodontitis related TNF-α -308 AA homozygous genotype was observed in one patient in GAP group with 1% frequency, whereas it was not observed in the control and CP groups. Homozygous AA and heterozygous GA genotypes were present 1 % and 47.6% of patients with GAP respectively, while frequencies were 0% and 11.8% in healthy control group respectively (p=0.000; Table 3). While the TNF-α -308 homozygous AA genotype was not present in both healthy controls and patients with CP, heterozygous GA genotype was present in 11.8% of controls and 31.9 % of patients with CP (p=0.001; Table 3). It is also observed that the frequency of polymorphic A allele was found in 84% of CP patients and 25% of GAP patients and this difference was statistically significant (p= 0.019).
| CP | GAP | Control | p value | p value | |||
|---|---|---|---|---|---|---|---|
| CP versus controls | GAP versus controls | ||||||
| TNF-α | Genotypes | GG | 81 (68.1%) | 54 (51.4%) | 75 (88.2%) | 0.001 | 0 |
| GA | 38 (31.9%) | 50 (47.6%) | 10 (11.8%) | ||||
| AA | 0(0%) | 1 (1%) | 0 (0%) | ||||
| n=119 | n=105 | n=85 | |||||
| Alleles | Allele G | 38(16%) | 158 (75%) | 160 (94%) | 0.002 | 0 | |
| Allele A | 200 (84%) | 52 (25%) | 10 (6%) | ||||
| n=238 | n=210 | n=170 | |||||
| LT-α | Genotypes | AA | 66 (55.5%) | 54 (51.4%) | 51 (60%) | 0.655 | 0.274 |
| AG | 45 (37.8%) | 45 (42.9%) | 27 (31.8%) | ||||
| GG | 8 (6.7%) | 6 (5.7%) | 7 (8.2%) | ||||
| n=119 | n=105 | n=85 | |||||
| Alleles | Allele A | 177 (74.4%) | 153 (73%) | 129 (76%) | 0.817 | 0.798 | |
| Allele G | 61 (25.6%) | 57 (27%) | 41 (24 %) | ||||
| n=238 | n=210 | n=170 | |||||
| TNFR-2 | Genotypes | TT | 10 (8.4%) | 9 (8.6%) | 40 (47.1%) | 0 | 0 |
| TG | 42 (35.3%) | 32 (30.5%) | 19 (22.4%) | ||||
| GG | 67 (56.3%) | 64 (61%) | 26 (30.6%) | ||||
| n=119 | n=105 | n=85 | |||||
| Alleles | Allele T | 62 (26.1%) | 50 (23.8%) | 99 (58.2%) | 0 | 0 | |
| Allele G | 176 (73.9%) | 160 (76.2%) | 71 (41.8%) | ||||
| n=238 | n=210 | n=170 | |||||
Table 3: Distribution of TNF-α -308, LT-α +252 and TNFR-2 +587 genotypes and allele frequencies for patients with CP and GAP and control group.
LT-α +252: Regarding LT-α +252 polymorphism, no statistically significant difference was found between the patients and the control subjects in Turkish population in this study (Table 3).
TNFR-2 (+587): For TNFR-2 (+587) T/G polymorphism; TT, TG and GG genotype and allele frequencies were analysed. TNFR-2 (+587) GG genotype was observed in 26 healthy subjects whereas in 64 GAP patients and in 67 CP patients (p= 0.000; Table 3). The frequencies of polymorphic G alleles were 73.9% in CP patients; 76.2% in GAP patients and 41.8% in controls (p= 0.000; Table 3).
The correlations between TNF- α -308, LT-α +252 and TNFR-2 (+587) polymorphisms and gender and the polymorphisms were investigated and no significant correlation was found in the groups (p>0.05).
Discussion
Periodontitis is a multifactorial disease triggered by the immune response to the products of pathogenic microorganisms and developed by the complex effects of environmental and genetic factors [15]. Genetic modifications, which can affect the inflammatory response of the host, are crucial to determine the susceptibility to periodontitis. These modifications may act as defence or risk factors changing the susceptibility and the severity of a disease [16]. Therefore, the pursuit of genetic markers related to the severity and the susceptibility for periodontitis has been receiving considerable attention. It is critical to find out the gene polymorphisms associated with periodontitis for diagnosis and treatment of the disease.
It is known that gene polymorphisms associated with immunity and inflammation can increase susceptibility and severity in diseases possessing inflammatory component like periodontitis [17]. TNF- α is one of the most important candidate genes related to periodontitis since it is a proinflammatory cytokine having role as a mediator. Besides, having functions parallel to disease factors such as fibroblast differentiation for forming periodontal ligaments and increasing the capacity of bone resorption is considered as a strong proof of TNF- α being a critical factor in the pathology of periodontitis [18]. It is stated that the expression level of TNF-α might be related to HLA-DR alleles or LT-α gene. A study conducted with adult periodontitis patients reported the relationship between NcoI polymorphism in the first intron of TNF-β (LT- α) gene and high level of TNF-α production and stated that this condition leads to susceptibility to inflammatory diseases [19].
The association between LT-α (+252) gene polymorphism and periodontitis has been investigated in various studies. However, in our study no statistically significant difference was found between CP-control, GAP-control and CPGAP groups for LT-α (+252) gene polymorphism. Therefore, it could be useful to increase the sample size to get more precise profile for Turkish population.
In our study; statistically significant differences were found between CP and control, and GAP and control groups in terms of TNF-α -308 GA genotypes and A allele. The prevalence of wild type TNF-α -308 GG genotype was significantly higher in the control group when compared to periodontitis (CP and GAP) groups. Therefore, it might be considered that this genotype is a protective factor in terms of periodontitis. Another important result obtained from our study is about TNF-α -308 GA genotype. In all groups, statistically significant differences were found for TNF-α -308 GA genotype. The presence of heterozygote genotype was lower in the control group, while it was higher in CP and GAP groups. It is crucial to state that the prevalence of the TNF-α -308 GA genotype was statistically significantly higher in generalized aggressive periodontitis which is characterized by more rapid disease development and severe attachment loss than chronic periodontitis. It can be concluded that; TNF-α -308 GA genotype may increase the incidence and severity of periodontitis and may predispose susceptibility to aggressive periodontitis in Turkish population. Susceptibility to periodontitis in the presence of TNF-α -308 GA genotype was also supported by the presence of A allele. The frequency of A allele was found to be statistically significant not only between the control and periodontitis groups but also between the CP and GAP groups. This is the first study reporting the association between the presence of TNF-α -308 GA genotype and A allele and the susceptibility to periodontitis. It is observed that only one A allele can increase the susceptibility to the disease. Therefore, it is suggested that TNF-α -308 GA genotype should be considered as a risk factor for periodontitis in Turkish population.
Third polymorphism which was investigated in the present study was TNFR-2, which is a surface receptor of TNF-α and has an active role in triggering inflammatory stages and pathogenesis of periodontitis. TNFR-2 gene polymorphisms were found to be associated with various periodontal diseases. Moreover, it was emphasized that the higher expression of TNFR- 2 could be used as a marker of periodontitis [20]. In our study; reflecting the Turkish population, an association between TNFR-2 (+587 T/G) gene polymorphism and periodontitis was determined. Statistically significant differences were observed between the, CP, GAP and control groups, in terms of genotypes and allele frequencies. The ratio of having polymorphic G allele was higher in periodontitis groups than the control group. It can be stated that TNFR-2 +587 GG genotype is associated with the incidence and the severity of periodontitis and that the rate is the highest in the GAP group in Turkish population. Furthermore; here we showed weak linkage disequilibrium between TNF- α and LT- α, TNF- α and TNFR-2, LT- α and TNFR-2 and concluded that no significant evidence of linkage disequilibrium was observed in the Turkish population. Smoking is considered as one of the risk factors of periodontitis [21]. When we checked the correlation between TNF-α -308 genotypes and smoking, TNFR-2 +587 genotypes and smoking and LT-α +252 genotype and smoking, no significant association was found.
Conclusion
It is possible to use TNF-α -308 GA genotype and TNFR-2 +587 GG genotype as a marker of susceptibility to periodontitis, if wider and more comprehensive studies are planned. Periodontitis is a multifactorial disease associated with pathogen, host, environmental and genetic factors. Therefore, conducting studies including more candidate genes, familial and haplotype analyses is crucial to determine the risk factors and protective strategies for periodontitis.
Conflict of Interest and Source of Funding Statement
The authors report no conflicts of interest related to this study.
References
- Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol 2003; 74:391–401.
- Kádár K, Kovács M, Karádi I, et al. Polymorphisms of TNF-alpha and LT-alpha genes in multiple myeloma. Leuk Res 2008; 32:1499–504.
- Loos BG, John RP, Laine ML. Identification of genetic risk factors for periodontitis and possible mechanisms of action. J Clin Periodontol 2005; 32:159–179.
- Yoshie H, Galicia JC, Kobayashi T, et al. Genetic polymorphisms and periodontitis. Int Congr Ser 2005; 1284:131–139.
- Boyce BF, Li P, Yao Z, et al. TNF-alpha and pathologic bone resorption. Keio J Med 2005; 54:127–131.
- Carswell EA, Old LJ, Kassel RL, et al. An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci USA 1975; 72:3666–3670.
- Goetz FW, Planas J V, MacKenzie S. Tumor necrosis factors. Dev Comp Immunol 2004; 28:487–497.
- Fassmann A, Holla LI, Buckova D, et al. Polymorphisms in the +252(A/G) lymphotoxin-alpha and the -308(A/G) tumor necrosis factor-alpha genes and susceptibility to chronic periodontitis in a Czech population. J Periodontal Res 2003; 38:394–399.
- Boraska V, Zeggini E, Groves CJ, et al. Family-based analysis of tumor necrosis factor and lymphotoxin-alpha tag polymorphisms with type 1 diabetes in the population of South Croatia. Hum Immunol 2009; 70:195–199.
- Fanning GC, Bunce M, Black CM, et al. Polymerase chain reaction haplotyping using 3’ mismatches in the forward and reverse primers: application to the biallelic polymorphisms of tumor necrosis factor and lymphotoxin alpha. Tissue Antigens 1997; 50:23–31.
- Zhang J, Sun X, Xiao L, et al. Gene polymorphisms and periodontitis. Periodontol 2000. 2011; 56:102–124.
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol 1999; 4:1–6.
- Löe H, Silness J. Periodontal disease in pregnancy I. Prevalence and severity. Acta Odontol Scand 1963; 21:533–551.
- Silness J, Löe H. Periodontal disease in pregnancy II. Correlation between oral hygiene and periodontal condition. Acta Odontol Scand 1964; 22:121–135.
- Bascones-Martínez A, Muñoz-Corcuera M, Noronha S, et al. Host defence mechanisms against bacterial aggression in periodontal disease: Basic mechanisms. Med Oral Patol Oral Cir Bucal 2009; 14:680–685.
- Craandijk J, van Krugten MV, Verweij CL, et al. Tumor necrosis factor-alpha gene polymorphisms in relation to periodontitis. J Clin Periodontol 2002; 29:28–34.
- Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol 2008; 79:1514–1519.
- Manolagas SC. Role of cytokines in bone resorption. Bone 1995; 17:63S-67S.
- Hollá LI, Fassmann A, Vašků A, et al. Interactions of Lymphotoxin α (TNF-β), angiotensin-converting enzyme (ACE), and endothelin-1 (ET-1) gene polymorphisms in adult periodontitis. J Periodontol 2001; 72:85–89.
- Morita C, Horiuchi T, Tsukamoto H, et al. Association of tumor necrosis factor receptor type II polymorphism 196R with systemic lupus erythematosus in the Japanese molecular and functional analysis. Arthritis Rheum 2001; 44:2819–2827.
- Haber J. Smoking is a major risk factor for periodontitis. Curr Opin Periodontol 1994; 12–18.
Author Info
Selin Özkan-Kotiloğlu1, Gülcan Kuyucuklu2, Esra Baltacioglu3*, Güliz Güncü4, Erhan Dursun4, Erkan Sukuroglu3, Erdem Karabulut5, Emine Sibel Sümer6 and Ferda Alev Akal?n4
1Department of Molecular Biology and Genetics, Faculty of Arts and Sciences, Ahi Evran University, Kırşehir, Turkey2Department of Medical Microbiology, Faculty of Medicine, Trakya University, Edirne, Turkey
3Department of Periodontology, Faculty of Dentistry, Karadeniz Technical University, Trabzon, Turkey
4Department of Periodontology, Faculty of Dentistry, Hacettepe University, Ankara, Turkey
5Department of Biostatistics, Faculty of Medicine, Hacettepe University, Ankara, Turkey
6Department of Biology, Faculty of Sciences, Hacettepe University, Ankara, Turkey
Citation: Selin Özkan-Kotilo?lu, Gülcan Kuyucuklu, Esra Baltacioglu, Güliz Güncü, Erhan Dursun, Erkan Sukuroglu, Erdem Karabulut, Emine Sibel Sümer, Ferda Alev Akal?n, Investigation of TNF-?, LT-? and TNFR-2 gene Polymorphisms in Patients with Periodontitis in Turkey, J Res Med Dent Sci, 2020, 8 (7): 173-179.
Received: 25-Sep-2020 Accepted: 28-Oct-2020 Published: 04-Nov-2020